All published articles of this journal are available on ScienceDirect.
Non-targeted Serum Metabolomics Identifies Candidate Biomarkers Panels Associated with Nonalcoholic Fatty Liver Disease: A Pilot Study in Russian Male Patients
Abstract
Aims:
The aim of the present study was to explore changes in the serum metabolome of patients with NAFLD relative to healthy controls to identify biomarkers associated with steatosis or Non-Alcoholic Steatohepatitis (NASH).
Background:
The serum metabolome reflects changes at the organismal level. This is especially important in Non-Alcoholic Liver Disease (NAFLD), where changes in hormones, cytokines, enzymes and other metabolic alterations can affect the liver, as well as adipose tissue, skeletal muscle and other systems.
Objective:
The objectives were to conduct non-targeted serum metabolomics, data processing, and identification of candidate biomarkers, as well as panels and assessment of their prognostic value.
Materials and Methods:
Non-targeted metabolomic analysis of blood serum samples from 21 male patients with NAFLD (simple steatosis or NASH) and seven male Control group was performed using gas chromatography-mass spectrometry.
Results:
A total of 319 serum metabolites were detected in NAFLD and Control groups, several of which differed significantly between groups. The most discriminating biomarkers were 3-hydroxybutyric acid, 2-hydroxybutyric acid, 2,3-dihydroxybutyric acid, arabitol and 3-methyl-2-oxovaleric acid. Using a panel of three, four or more markers could distinguish patients with NAFLD from controls, and patients with NASH from those with simple steatosis.
Conclusion:
We identified candidate biomarkers for simple steatosis and NASH. Since NAFLD is a multifactorial disease, it is preferable to use a marker panel rather than individual metabolites. Markers may not only result from dysregulation of metabolic pathways in patients with NAFLD, they may also reflect adaptive responses to disease, including functional changes in the intestinal microbiota.
1. INTRODUCTION
Chronic Liver Disease (CLD), and especially Non-Alcoholic Fatty Liver Disease (NAFLD), is becoming an increasing burden worldwide, both medically and financially. The global prevalence of NAFLD is ~25% [1]. Since 60-75% of CLD patients are predicted to suffer from NAFLD, the total number of patients with CLD may be in the range of 2.5-3 billion people worldwide.
NAFLD is associated with obesity, insulin resistance, type 2 diabetes mellitus, arterial hypertension, dyslipidaemia, and metabolic syndrome. NAFLD stages range from Simple Steatosis (SS) to Non-Alcoholic Steatohepatitis (NASH), which can progress to fibrosis and cirrhosis, Hepatocellular Carcinoma (HCC), and liver transplantation. NASH is an important cause of liver cirrhosis. From 1990 to 2017, the prevalence of compensated cirrhosis due to NASH more than doubled, and cases of decompensated cirrhosis more than tripled due to NASH [2].
Until recently, the ‘two hits’ theory was routinely applied to NAFLD pathogenesis, in which the first hit is steatosis development, and the second hit is steatohepatitis. This concept is now outdated and has been replaced by the ‘multiple hits’ hypothesis, which more accurately reflects the complex mechanisms triggering the onset and progression of NAFLD. This concept includes pathogenetic factors such as insulin resistance, adipose tissue hormones, obesity, diet, genetic and epigenetic factors, as well as the gut-liver axis, which appears to play a key role in the development and progression of NAFLD. The leading players in this axis are the gut microbiota, bacterial metabolites and intestinal barrier [3].
Early diagnosis of NAFLD and monitoring of disease progression via metabolomics is extremely important. Metabolites determine the molecular phenotype of an organism since they are the substrates, intermediates and products of biochemical reactions. Therefore, changes are reflected in the metabolome, including those associated with the progression of pathological processes. In particular, serum metabolome analysis provides an opportunity for efficient diagnosis of various diseases [4]. Technological advances in recent years make it possible to identify hundreds or even thousands of metabolites in a single sample in just a few minutes, which is ideal for the diagnosis of multifactorial diseases such as NAFLD [5, 6].
In the present study, we compared the serum metabolomes of individuals with simple steatosis or NASH with those of controls. Together with the accumulating literature, our results indicate that markers may not only reflect dysregulation of metabolic pathways, but also adaptive responses to disease, and they may indicate regulatory effects on organisms [7-11].
2. MATERIALS AND METHODS
2.1. Subjects
The subjects in this study comprised 28 males aged 49 ± 5 years. Only male patients were included because NAFLD is a sexual dimorphic disease, and high levels of oestrogen can protect against NAFLD development and progression [12]. In addition, there are gender-specific differences in the human metabolome; about a third of serum metabolites differ significantly between males and females [13, 14]. The subjects included seven male controls, 10 patients with simple steatosis (SS) and 11 with NASH. Patients were recruited from North-Western State Medical University named after I.I. Mechnikov. Diagnosis of NAFLD was confirmed by anamnesis data, laboratory and instrumental research methods, non-invasive tools (FibroMax [BioPredictive, Paris, France]) and liver biopsies. Exclusion criteria included chronic viral hepatitis, autoimmune-, alcoholic-, drug-induced, and genetic-related liver disease. A fasting blood sample was obtained from subjects in the morning, serum was separated by centrifugation and stored at - 40°C until analysis.
2.2. Ethics
The study protocol was approved by the Ethics Boards of North-Western State Medical University named after I.I. Mechnikov (Protocol Nº7), and it conformed to the ethical guidelines of the 1975 Declaration of Helsinki. All participants gave their informed written consent. All methods were performed in accordance with the relevant guidelines and regulations of North-Western State Medical University and State Research Institute of Highly Pure Biopreparations.
2.3. Sample Preparation
Samples were thawed at room temperature and metabolites were extracted with acetonitrile (Cryochrome, Russia) with simultaneous precipitation of proteins. A 0.1 ml volume of serum was removed from chilled samples, added to 0.5 ml of acetonitrile, vortexed for 3 min, centrifuged at 13,000 rpm for 3 min, and the clear supernatant was collected. The resulting extract was dried under a stream of nitrogen until a dry residue was obtained. An internal standard, tridecanoic acid trideuteromethyl ester (2 mg/ml or 8.7 mM; FisherSci, USA) dissolved in methanol, was added to the dry residue and dried again. Derivatives were prepared by silylation using N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA; Supelco, USA).
2.4. GC-MS Analysis
Gas Chromatography-Mass Spectrometry (GC-MS) analysis was carried out using a GCMS-QP2010 Plus instrument (Shimadzu, Japan) equipped with an Agilent HP Ultra-2 analytical capillary column containing (5%-phenyl)-methylpolysiloxane resin (25 m length, 0.2 mm inner diameter, 0.25 μm stationary phase film thickness). The column was heated from 50 to 290°C at 10oC/min. The volume of the injected sample was 100 μl, the flow division was 1:50, the carrier gas (helium) flow rate was 1 ml/min, and the injector and detector temperature was 280°C. The chromatogram was recorded in two modes; (1) from 1 min to 6.4 min, monitoring of ions m/z 103, 117 and 145 (Selected Ion Monitoring (SIM) mode); (2) from 6.4 min to 40 min, total ion current monitoring in the mass range 35 to 550 (scan mode). Each chromatogram was obtained by recording the total ion current at a frequency of 2.5 scans/s.
2.5. Data Processing
Raw datasets were acquired using GCMS Analysis software (GCMS solution, Shimadzu, Japan). Firstly, was used g MetAlign data pre-processing tool (www.wur.nnl/Onderzoek -Resultaten/Onderzoeksinstituten/food-safety-research/show-rikilt/MetAlign.htm), followed by AIoutput software (www.prime.psc.riken.jp/Metabolomics_Software/AIoutput/index.html), which can perform the peak identification, prediction, and data integration from the result exported from MetAlign. Next, peak detection, deconvolution and identification according to retention index (RI), retention time (RT) and mass spectra were performed using GCMS solution and Automated Mass Spectrometry Deconvolution and Identification System software (AMDIS, www.amdis.net, version 2.73) with the National Institute of Standards and Technology (NIST) database. Additionally, identification of compounds was performed using the Human metabolome database (HMDB, www.hmdb.ca) and the GOLM metabolome database (GOLM, www.gmd.mpimp-golm.mpg.de). To confirm compounds, at least three characteristic ions were used. Mass spectra were considered annotated when they matched the library variant with a match factor ≥ 80. The amount of compound in each sample was calculated using the internal area normalisation method. The peak area of the total ionic current of a compound was divided by the peak area of the internal standard, and the obtained value was the normalised amount of compound in the sample.
2.6. Statistical Analysis
Differences in the clinical parameters between the control group and patients were tested using Student’s t-test. Significance was defined as p<0.05. Metabolomics data were assessed by Principal Component Analysis (PCA). The search for biomarkers that can distinguish between different groups was carried out using multivariate analysis, comprising partial least squares discriminant analysis (PLS-DA), Support Vector Machine (SVM) analysis, and a Naïve Bayes simple probabilistic classifier. The results of experiments were used to construct a table containing the accuracy and variance for the compared methods and predicted uncertainty matrices. As a result, a Receiver Operating Characteristic (ROC) curve was plotted for each classifier, and classifiers with the highest sensitivity and specificity (AUC) values were chosen. Each classifier ranked all compounds in descending order of importance. From this list, the first 30 compounds contributing most to the differences between the two groups were selected. Mann-Whitney tests were performed to compare data obtained from experimental groups, and p <0.05 was considered significant. Statistical analysis of the data was performed using the freely available R software package (http://cran.r- project.org/). For each candidate biomarker, ROC curves were plotted and AUC values were determined. Selecting the optimal combination of biomarkers is a complex process and requires the integration of data signatures using advanced statistical techniques. To select the biomarker panels, CombiROC software was employed (www.combiroc.eu).
3. RESULTS
The study involved male patients with NAFLD, and controls with non-pathological liver ultrasound patterns, normal non-invasive blood test (FibroMax) results, and normal Alanine Transaminase (ALT) and Aspartate Transaminase (AST) levels (Table 1). Body Mass Index (BMI) was 25-30 kg/m2 for controls, and 30-35 kg/m2 (class I obesity) for patients with NAFLD. Patients were characterised by increased ALT and AST levels and significant steatosis (progressive stages of fibrosis were observed in some patients with NASH).
| Indicator | Controls (n = 7) |
Simple Steatosis (n = 10) |
NASH (n = 11) |
|---|---|---|---|
| Age (years) | 49.8 ± 6.1 | 49.3 ± 5.4 | 48.4 ± 6.1 |
| BMI (kg/m2) | 27.6 ± 0.6 | 31.0 ± 2.5a | 32.6 ± 1.9a |
| AST (U/l) | 20.1 ± 8.2 | 34 ± 11.2b | 64.3 ± 31.2a, b |
| ALT (U/l) | 15.8 ± 7.4 | 60.1 ± 38.8a, b | 104.4 ± 61.7 a, b |
| Steatosis degree 0/1/2/3 | 7/0/0/0 | 0/2/4/4 | 0/0/4/7 |
| Fibrosis degree 0/1/2/3 | 7/0/0/0 | 4/6/0/0 | 0/2/7/2 |
b Significant at p<0.05 SS group vs. NASH group.
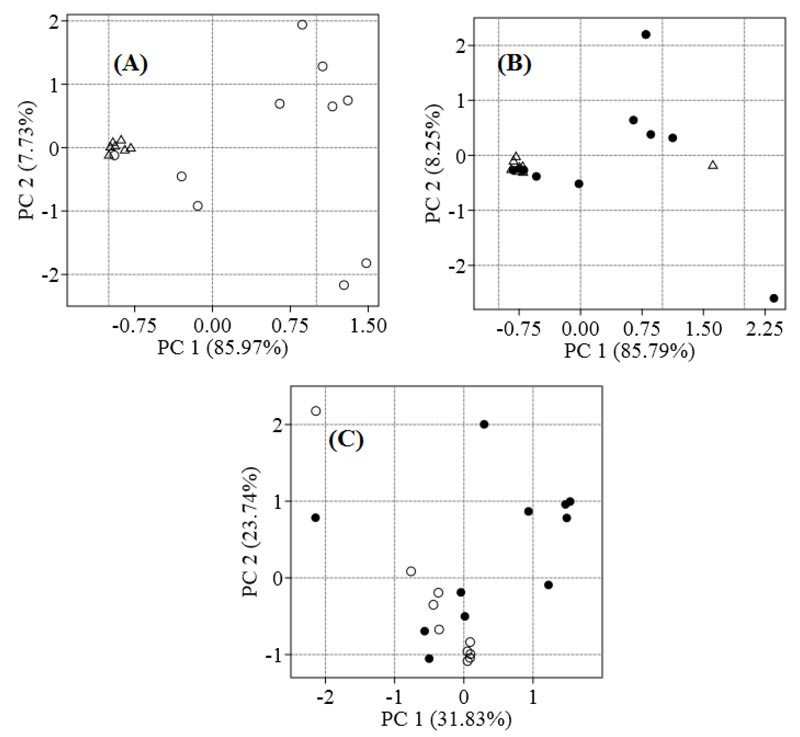
We identified 319 metabolites in patients with NAFLD and controls. To visualise the differences between the metabolomes of patients and controls, PCA was employed. Control samples were grouped on the left side of the plane relative to the axis of Principal Component 1 (PC1), and SS samples are distributed at different distances from each other, mainly along the right side of the plane (Fig. 1A). The seven NASH samples are located at a distance from controls, and the four samples overlap (Fig. 1B). The SS and NASH samples are spread out over the plane, and the steatosis group is different from the NASH group (Fig. 1C).
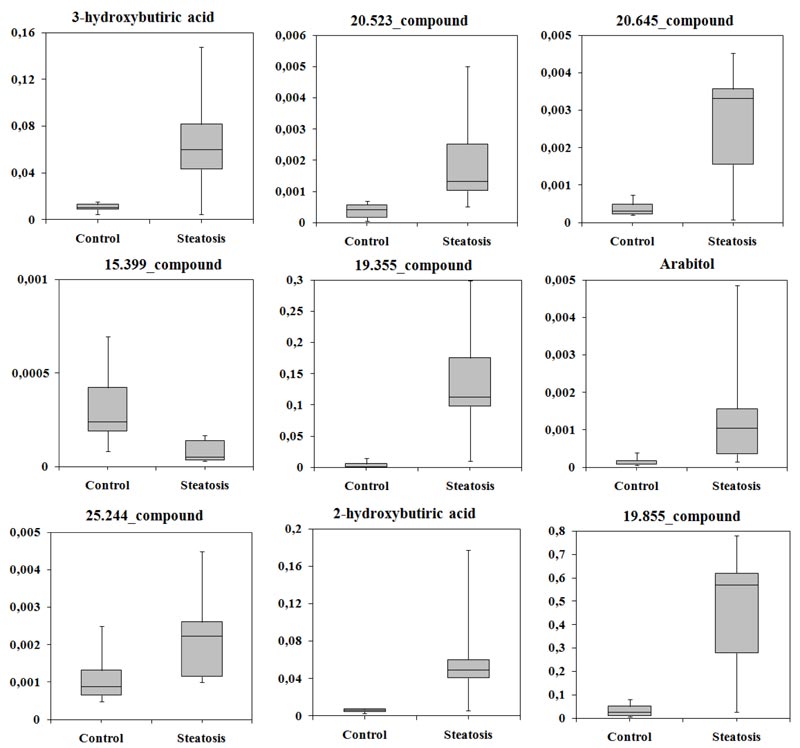
Compared with controls, the metabolomes of patients with SS were characterised by a significant increase in the amount of hydroxy acids: 2-hydroxybutyric acid (8.1-fold), 3-hydroxybutyric acid (β-OHB, a ketone body, 5.8-fold), 2-hydroxy-3-methyl butyric acid (3.4-fold); amino acids: threonine (7.5-fold), proline (5.3-fold), pyroglutamic acid (4.9-fold), and isoleucine, amino acid with a branched side chain (6.0-fold); sugar D-xylose (14.7-fold) and sugar alcohols arabitol (12.3-fold); and a number of unidentified compounds (Supplementary Material-1). Patients with NASH showed significantly higher serum levels of 3-methyl-2-oxovaleric acid (6.8-fold increase) and another four unidentified compounds than those with SS. A significant increase in levels of 2,3-dihydroxybutyric acid (7.3-fold), arabitol (17.9-fold), and another five compounds was observed in serum from NASH patients relative to controls.
Candidate biomarkers were identified using multivariate analysis (SVM, PLS-DA and Naïve Bayes). The classifier was chosen based on the largest area under the ROC curve of sensitivity and specificity. When comparing patients with SS and controls, the classifier was SVM (AUC = 0.961). The average accuracy of the classifier was 0.87, the variance of the accuracy was 0.026, the sensitivity was 88.2%, and the specificity was 83.2%. Based on SVM analysis, all compounds were sorted in decreasing order of importance. The first 30 compounds were selected for further statistical analysis, and nine of these compounds differed significantly (p < 0.05) between patient and control groups (Table 2).
| Compound Name or RT | Characteristic Ions |
Fold Change* (SS/Controls) |
p-value (Mann-Whitney U Test) |
Metabolic Pathways |
|---|---|---|---|---|
| 3-hydroxybutyric acid | 87;99;101 | 5.8 | 0.003 | Fatty acid biosynthesis |
| 20.523_compound | 103;117;161 | 3.2 | 0.003 | - |
| 20.645_compound | 191;204;217 | 11.1 | 0.005 | - |
| 15.399_compound | 156;172;216 | 0.21 | 0.0007 | - |
| 19.355_compound | 129;147;157 | 106.6 | 0.0004 | - |
| Arabitol | 103;117;129 | 12.3 | 0.002 | Interconversion of pentose and glucoronate |
| 25.244_compound | 95;103;129 | 2.5 | 0.013 | - |
| 2-hydroxybutyric acid | 87;95;101 | 8.1 | 0.001 | Metabolism of cysteine, methionine, S-adenosylmethionine and taurine |
| 19.855_compound | 133;147;189 | 21.6 | 0.007 | - |
RT, retention time (min); *, the ratio of the medians.
Some compounds were not annotated, but their retention time (RT), a set of characteristic ions, and their mass spectra could still be used. In patients with SS, levels of eight compounds were increased, while 15.399_compound was decreased approximately five-fold compared with controls. 3-hydroxybutyric acid (β-OHB), 2-hydroxybutyric acid, and arabitol were identified as candidate biomarkers. The normalised levels of candidate biomarkers in serum are shown in (Fig. 2). Notably, levels of these compounds differ more between patients in the SS group than in controls.
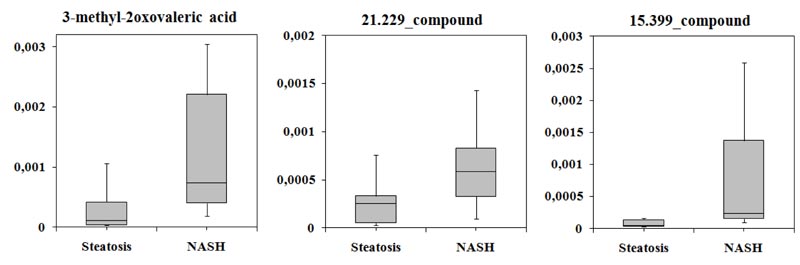
Next, was identified candidate biomarkers that could distinguish patients with SS from those with NASH. Analysis of ROC curves for three classifiers showed that the largest area under the curve corresponded to PLS-DA (AUC = 0.992). The average accuracy of the classifier was 0.985, the variance of the accuracy was 0.0035, and sensitivity and specificity were 98.5%. It was shown levels of 3-methyl-2-oxovaleric acid, 21.229_compound, and 15.399_compound differed significantly between the two groups (Table 3).
Levels of all compounds increased by more than two-fold in the serum of patients with NASH compared with the SS group and varied greatly (Fig. 3).
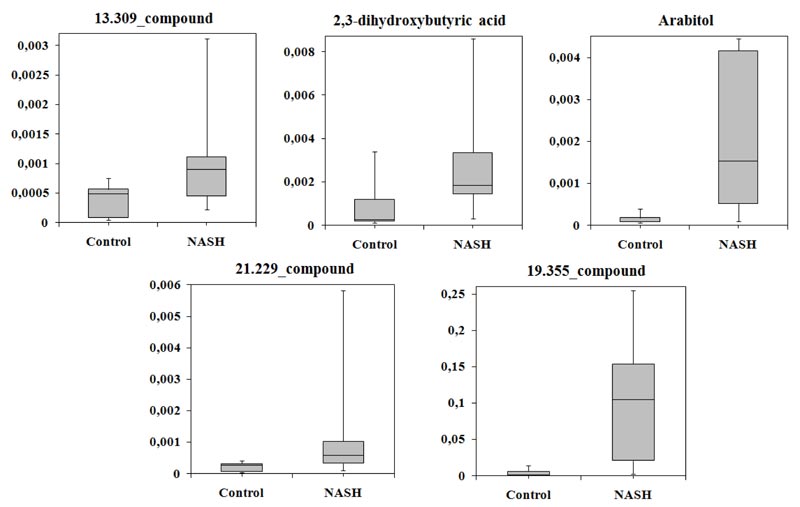
Multivariate analysis of the metabolomes of NASH patients and controls based on three classifiers showed that the largest area under the ROC curve corresponded to SVM (AUC = 0.788). The average accuracy of the classifier was 0.81, the variance of the accuracy was 0.027, the sensitivity was 100%, and the specificity was 63.7%. Following SVM analysis, 2,3-dihydroxybutyric acid, arabitol, 13.309_compound, 21.229 _compound, and 19.355_compound differed significantly between the groups, and were selected. Levels of all these compounds were increased in patients with NASH (Table 4).
| Compound Name or RT | Characteristic Ions |
Fold Change (NASH)/(SS) |
p-value (Mann-Whitney U Test) |
Metabolic Pathways |
|---|---|---|---|---|
| 3-methyl-2oxovaleric acid | 85;101;113 | 6.8 | 0.010 | Metabolism of valine, leucine and isoleucine |
| 21.229_compound | 73;147;156 | 2.3 | 0.029 | - |
| 15.399_compound | 156;204;217 | 4.7 | 0.0008 | - |
| Compound Name or RT | Characteristic Ions |
Fold Change (NASH)/(Controls) |
P-value (Mann-Whitney U Test) |
Metabolic Pathways |
|---|---|---|---|---|
| 13.309_compound | 133;147;175 | 1.9 | 0.04 | - |
| 2,3-dihydroxybutyric acid | 57; 73; 103 | 7.3 | 0.027 | - |
| Arabitol | 103;117;129 | 17.9 | 0.0008 | Interconversion of pentose and glucoronate |
| 21.229_ compound | 73;147;156 | 2.2 | 0.02 | - |
| 19.355_ compound | 129;147;157 | 99.0 | 0.0008 | - |
The level of all compounds more than 50% increased in serum of the patients with NASH. As in previous comparisons, the normalized level of the candidate biomarkers varied greatly in NASH group (Fig. 4).
| Compound Name or RT | AUROC | Sensitivity | Specificity | CI 95% |
|---|---|---|---|---|
| SS vs. Controls | ||||
| 3-hydroxybutyric acid | 0.914 | 1 | 0.9 | 0.744-1 |
| Arabitol | 0.929 | 0.714 | 1 | 0.809-1 |
| 2-hydroxybutyric acid | 0.943 | 1 | 0.9 | 0.8241-1 |
| 19.355_compound | 0.971 | 1 | 0.9 | 0.9048-1 |
| 20.523_compound | 0.914 | 1 | 0.8 | 0.7771-1 |
| 20.645_compound | 0.900 | 1 | 0.9 | 0.704-1 |
| 15.399_compound | 0.957 | 0.857 | 1 | 0.8629-1 |
| 25.244_compound | 0.857 | 0.714 | 1 | 0.6535-1 |
| 19.855_compound | 0.957 | 1 | 0.9 | 0.8641-1 |
| NASH vs. SS | ||||
| 3-methyl-2oxovaleric acid | 0.827 | 0.6 | 1 | 0.644-1 |
| 21.229_compound | 0.782 | 0.7 | 0.818 | 0.578-1 |
| 15.399_compound | 0.909 | 0.7 | 1 | 0.786-1 |
| NASH vs. Controls | ||||
| 2,3-dihydroxybutyric acid | 0.818 | 0.714 | 1 | 0.5727-1 |
| Arabitol | 0.948 | 1 | 0.818 | 0.8537-1 |
| 21.229_compound | 0.831 | 1 | 0.545 | 0.6387-1 |
| 13.309_compound | 0.792 | 1 | 0.545 | 0.5752-1 |
| 19.355_compound | 0.948 | 1 | 0.818 | 0.8533-1 |
ROC curves were constructed, and the Area Under the Curve (AUC) was calculated for each candidate biomarker. The area under the ROC curve ranged from 0.782 to 0.971 (Table 5).
Finally, we analysed combinations of biomarkers. In most cases, panels yielded higher AUROC (0.998), sensitivity (1), and specificity (1) values than individual biomarkers. Paired combinations with candidate biomarkers 19.355_compound and 20.523_compound achieved the highest prognostic indicator values for SS and Control groups. The predictive value of panels of three, four or more candidate biomarkers also yielded the highest values (AUROC = 0.998, sensitivity = 1, specificity = 1). The panel including 3-methyl-2-oxovaleric acid, 15.399_compound, and 21.229_compound achieved the highest prognostic value for distinguishing SS and NASH patients. The panel with 21.229_compound and arabitol, as well as panels of three, four and five candidate biomarkers achieved the highest AUROC (0.998), sensitivity (1), and specificity (1) values for distinguishing NASH patients from controls.
4. DISCUSSION
Pathological processes lead to changes in specific metabolic pathways, which is in turn reflected by changes in serum metabolites (i.e. the serum metabolome). Thus, metabolites are not only indicators of the dysregulation of metabolic pathways but factors of pathogenesis and/or the responses to a pathological state.
In the present study, we compared the serum metabolomes of patients with NAFLD and controls with non-pathological liver. The software packages used for data processing identified 319 compounds and 108 compounds were previously annotated. Non-annotated compounds were characterised by retention time and characteristic ions. The distribution of samples from patients and controls obtained by PCA revealed differences in metabolomic profiles between patients with SS and NASH, and between both patient and control groups. Conversely, similarities between groups may indicate similarity between metabolomic profiles.
The metabolome analysis results revealed significant changes in certain key pathways, specifically glutathione metabolism, and lipid and amino acid metabolism (Supplementary Material 1). Some of the identified metabolites for which levels varied significantly between groups (isoleucine, proline, 3-hydroxybutiric acid, arabitol, 3-methyl-2-oxovaleric acid, 2-hydroxy-3-methylbutyric acid) were related to endogenous and/or microbial production. For example, the increase in 2-hydroxy-3-methylbutyric acid (2-hydroxyisovaleric acid) in patients with SS could be the result of increased production by Proteus mirabilis, Eggerthella lenta, or Listeria spp., as well as chronic intestinal inflammation [15]. Among the amino acids was identified isoleucine (6-fold increased in patients with SS compared to controls, p = 0.025), a member of the branched-chain amino acid (BCAA) group, which has been associated with an increased risk of metabolic disease, including insulin resistance (IR) and NAFLD [16, 17]. The level of pyroglutamic acid (or 5-oxoproline) in patients with SS was 5-fold higher than controls (p = 0.01). Another previous study demonstrated the high diagnostic value of pyroglutamic acid, for separating patients with steatosis from patients with NASH [18]. The concentration of pyroglutamate in serum of patients with steatosis was increased 1.56-fold compared with the control group, and 2.26-fold in patients with NASH compared with steatosis. In our current work, the results of multivariate analysis indicated that this compound was not a biomarker. In addition, an increase in the level D-xylose (14.7-fold) was observed in serum, which is presumably associated with increased intestinal permeability in disease patients. A similar explanation appears to be applicable for the increased serum level 1-kestose in serum from NASH patients.
Multivariate analysis of the metabolomes of patients with SS and controls showed that the levels of nine candidate biomarkers differed significantly between SS and control groups. The SS patients displayed a 5.8-fold increase in 3-hydroxybutyric acid (β-hydroxybutyrate, β-OHB), a major ketone body. Most ketone bodies are produced in the liver [19], although small amounts can be produced in other tissues through the aberrant expression of ketogenic enzymes or alteration of the ketolysis pathway. The observed increase in 3-hydroxybutyric acid is probably associated with an increase in beta-oxidation, as well as an increase in oxidative metabolism in the liver in general. Increased levels of this metabolite in patients with SS were reported previously. Interestingly, the authors reported a decrease in 3-hydroxybutyric acid with the progression from SS to NASH [20]. Presumably, an increase in β-OHB is an adaptive response that protects the liver against NAFLD progression during the early stages of SS. Subsequently, the progression of NAFLD leads to impaired ketogenesis and the development of maladaptive ketogenic insufficiency, contributing to NASH and hyperglyacemia. Therefore, levels of β-OHB in NASH may decrease [8, 9].
The second most important biomarker was 2-hydroxybutyric acid, level of which was increased 8.1-fold in patients. This compound is derived from α-ketobutyric acid, which is formed mainly in the liver during the catabolism of L-threonine, and the synthesis of glutathione [21]. Oxidative stress or enhanced detoxification of xenobiotics in the liver stimulates a sharp increase in the rate of glutathione synthesis, which can lead to increased production of 2-hydroxybutyric acid as an intermediate metabolic product [22, 23]. Recent studies have shown that an increased concentration of 2-hydroxybutyric acid may reflect early signs of IR, and serve as an independent predictor of the development of impaired glucose tolerance (prediabetes) and early-stage type 2 diabetes [22, 24-27].
In the SS group, the sugar alcohol arabitol was increased 12.3-fold relative to controls. Sugar alcohols are hydrogenated forms of carbohydrates in which the carbonyl group (aldehyde or ketone-reducing sugar) has been reduced to a primary or secondary hydroxyl group. Increased levels of arabitol in plasma and urine have been reported earlier for patients with congenital cirrhosis of the liver due to transaldolase deficiency [28].
It should be emphasised that we observed a high degree of variability in the amounts of the identified markers in patient serum. Since the serum metabolome reflects all changes in tissues and organs, and not just any individual organ, this may be especially important for NAFLD. Changes in hormones, cytokines, enzymes and other metabolic alterations can affect not only the liver, but also adipose tissue, skeletal muscle, and other systems. Thus, the observed variability may be due to differences in the state of organs and systems in different patients or to changes in metabolite levels over time.
Three candidate biomarkers distinguishing the SS and NASH groups were identified: 3-methyl-2-oxovaleric acid, 21.229_compound, and 15.399_compound. The most significant among them was 3-methyl-2-oxovaleric acid, which increased 6.8-fold with the progression of NAFLD. This metabolite is the first product of isoleucine degradation, and an increase in its concentration in serum indicates an increase in BCAA degradation. Another study demonstrated a correlation between the level of 3-methyl-2-oxovaleric acid and the development of type 2 diabetes mellitus [29]. A recent pilot study of the effects of curcumin on the serum metabolomic profile of patients with NAFLD showed that certain BCAA degradation products, such as 3-methyl-2-oxovaleric acid and 3-hydroxyisobutyrate, could consider both biomarkers and therapeutic targets for NAFLD [30]. It is possible that an increase in the level of this acid is associated not only with the increased degradation of BCAAs, but also with their production by the microbiota. Thus, in NAFLD, the amount of isoleucine produced by bacteria (Bacteroides vulgatus, Prevotella copri, Streptococcus sp., Clostridium sp., Eubacterium rectale) is increased [16, 31, 32]. Additionally, the body’s responses to a pathological process can alter metabolite levels. For example, when steatosis progresses to steatohepatitis, levels of BCAA: leucine (127%), isoleucine (139%) and valine (147%) are increased, while leucine supplementation activates the target of rapamycin (mTOR), which is a critical mediator regulating protein synthesis, cell proliferation and insulin sensitivity [33, 34]. In addition, BCAAs exert protective inhibition in cancer development. Thus, they may be increased under certain conditions, and their degradation products can be considered an adaptive response of the liver to oxidative stress during the NASH stage [35].
Multivariate analysis of NASH patient and control metabolomes allowed us to identify five candidate biomarkers. Their levels were increased from 1.9- (compound 13.309_
compound) to 99-fold (compound 19.355_compound) in the serum of patients with NASH. The second and third most important biomarkers were 2,3-dihydroxybutyric acid and arabitol; their levels were increased by 7.3- and 17.3-fold, respectively. 2,3-Dihydroxybutyric acid has two enantiomers: 4-deoxyerythronic acid ((2R,3R)-2,3-dihydroxybutanoic acid) and 4-deoxythreonic acid ((2S,3R)-2,3-dihydroxybutanoic acid). Currently, very little is known about the pathways of the formation of these entiomers. It is assumed that the main source of 4-deoxyerythronic acid is threonine. It has been found to be inversely associated with age in adults [36], and higher levels of 4-deoxyerythronic acid, 4-deoxythreonic acid, and 2-hydroxybutyric acid have been observed in children with type I diabetes [37]. 4-Deoxyerythronic acid is presumably formed from threonine by the action of threonine dehydrogenase, which is a relatively minor contributor to threonine oxidation in humans (about 10%) [38]. Lau C.E.
et al. suggest that an increased concentration of 4-deoxyerythronic acid may result not only from endogenous catabolism of threonine but also from exogenous sources or microbial metabolism [39]. Since it is assumed that 2,3-dihydroxybutyric acid may contribute to the pathophysiology of metabolic disorders such as obesity and diabetes [37, 39], we hypothesize its possible involvement in the pathogenesis of NAFLD.
All candidate biomarkers yielded good or excellent test results, but did not always have high sensitivity and specificity (Table 5). Panels of three to eight biomarkers yielded excellent test results with high sensitivity and specificity, thus it is preferable for diagnosis over the use of individual biomarkers. Panels of biomarkers have been successfully applied for the diagnosis of cancer, Parkinson’s disease, type 2 diabetes, and other multifactorial diseases [40-42]. NAFLD is a multifactorial disease, and given the various pathophysiological processes involved in the progression of NAFLD, it is doubtful whether a single marker could reflect all pathological changes. By contrast, a panel of markers can reflect the actual pathophysiological status of a patient, resulting in a more accurate diagnosis. In addition, the use of a panel of biomarkers obviously reduces the risk of misdiagnosis due to incorrect identification of a marker, or an incorrect measurement of its concentration in blood and also allows the inclusion of non-annotated compounds.
CONCLUSION
In conclusion, we identified nine biomarkers of SS, five biomarkers of NASH, and three biomarkers that distinguished SS from NASH patients. Since NAFLD is a multifactorial disease, we suggest that the use of a panel of markers is preferred over individual metabolites. We believe that markers may not only be the result of dysregulation of metabolic pathways in patients with NAFLD, but may also play a role in adaptive responses to disease and may therefore reflect functional changes in the intestinal microbiota. Further studies with a larger population are needed to confirm our hypotheses, and identify non-annotated biomarkers.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All procedures performed in the study were in accordance with the ethical standards and approved by the local ethical and deontology committee of North-Western State Medical University named after I.I. Mechnikov, St. Petersburg, Russia (Protocol No. 7).
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures followed were in conformity with the ethical standards of the committees responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was obtained from all subjects prior to the study.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
This work was financially supported by a grant from the President of the Russian Federation (Grant number MK-2429.2020.4, agreement No. 075-11-2020-007, dated July 22, 2020).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
We acknowledge the support for PhD students from the Department of Internal Diseases, Gastroenterology and Dietetics, North-Western State Medical University named after I.I. Mechnikov Abatsieva M.P., and help with sample preparation from senior researchers at the Department of Microbiology, State Research Institute of Highly Pure Biopreparations, Schalaeva O.N.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website, alongside the published article.


