All published articles of this journal are available on ScienceDirect.
Prognostic Value of Long Non-coding RNAs in Urological Malignancies: A Systematic Review, and Meta-Analysis
Abstract
Background:
Many studies have explored the potential roles of long non-coding RNAs (lncRNAs) in urological cancer (UC) progression. The clinical outcome and prognosis of UCs remain weak. Therefore, finding clinical prognostic markers is needed to improve therapeutic potential. The aim of this study was to consider the possible association between the lncRNAs expression with the survival time and clinical outcomes in patients with UC.
Methods:
A literature search was performed in several related databases to find eligible English papers published before 9 February 2021. Hazard ratios (HRs) with 95% CI were calculated to investigate the association between lncRNAs expression and overall survival in patients with UC.
Results:
A total of 46 studies, including 39 lncRNAs were identified. Results indicated that lncRNAs expression was significantly correlated with poor overall survival (OS) outcome in patients with UCs (HR: 1.923, 95% CI: 1.448-2.554, P<0.001). Also, we divided included studies into up-regulated and down-regulated subgroups according to lncRNAs expression. The results indicated a significant association with poor OS outcomes in both up-regulated (HR=2.546, 95% CI: 1.896-3.41, P<0.001) and down-regulated (HR=0.33, 95% CI: 0.22-0.49, P<0.001). Moreover, expression of lncRNAs was significantly associated with lymph node metastasis (LNM) (OR=0.25, 95% CI: 0.13-0.47, P<0.001)
Conclusion:
Abnormal expression of various lncRNAs is a potential novel marker for predicting the clinical outcomes of urological tumors.
1. INTRODUCTION
Urological Cancers (UCs) comprise approximately 1.5 percent of all malignancies and 12% of cancer-related deaths around the world. The incidences of UCs are the most common in the western world compared to Asian countries [1]. Prostate cancer, renal cell carcinoma, bladder, and testis are most commonly defined as urological malignancies. Among all tumor types related to the urological system, Prostate Cancer (PC) is the most prevalent and the third most common cause of cancer death in men [2]. The prevalence increases with age and these malignancies are the main source of morbidity and mortality in men over forty years old [3]. Despite the development of treatment for UCs, clinical outcome and prognosis remain weak and useful biomarkers to estimate prognosis and early diagnosis can increase the survival time and help optimal treatment of patients.
The rapid evolution of techniques for whole-genome and transcriptome sequencing have helped demonstrate the critical roles of long non-coding RNA (lncRNA) in the pathogenesis of a wide range of human diseases, especially cancer and tumor genesis [4, 5]. LncRNAs are a class of non-coding RNAs >200 nucleotides long but with no protein-coding potential that modulate different signaling pathways by acting as transcriptional or post-transcriptional regulators [6, 7]. There is accumulating evidence that they have a critical role in urological system malignancies, and these molecules have the potential to be used as biomarkers for diagnosis and prognosis in the clinic [8]. For example, Chen et al. indicated that up-regulated lncRNA-n336928 in patients with Bladder Cancer (BC) might have a relative with poor prognosis [9]. In another study, Xue et al. also demonstrated that NBAT1, as a novel lncRNA associated with poor prognosis in patients with cell renal cell carcinoma [10].
Currently, the special expression profiles of lncRNAs have been determined in UCs, and abnormal expression of lncRNAs that may regulate tumor genesis and metastasis-associated genes has been reported so as to discover the association between lncRNAs deregulation and estimation of survival rate and prognosis of patients with UCs; we conducted this quantitative meta-analysis.
2. METHODS
2.1. Search Strategy
A primary search was performed on the PubMed, Scopus, Web of Science, and Google scholar databases to find eligible English papers published before 9 February 2021. Also, we screened all selected paper references lists for further prospective periodicals. The search terms used included “Long Noncoding RNA” OR “lncRNA” OR “Long ncRNA” OR “Long Non-Translated RNA” OR “Neoplasm” OR “Cancer” OR “Malignancy” OR “Carcinoma” OR “Urogenital Neoplasm” OR “Genitourinary Neoplasm” OR “Genitourinary Cancers” OR “Urological Neoplasm” OR “Urological cancers” OR “Urinary Tract neoplasm” OR “Urinary Tract Cancers” OR “Urological malignancies” OR “Penis Cancers” OR ” Prostate Cancer” OR ” Testis Cancers” OR ” Renal Cancer” OR “Renal cell carcinoma” OR “Bladder cancer” OR ” Cancer of Ureter” OR “Prognosis” OR “Prognostic” OR “Survival” OR 'Outcome”. We did not try to assess not published papers.
2.2. Inclusion and Exclusion Criteria
The following criteria in this Meta-analysis were: 1) English papers, 2) studies that evaluated the lncRNA expression level in tissue samples with presented the clinicopathological features, 3) papers that explored the link between the expression level of lncRNA and survival rate, 4) papers that provided a Hazard ratio (HR) with 95% confidence interval (CI) or Kaplan Meier curves or sufficient data to extract HR and their 95% CI.
Exclusion criteria in this study included: 1) Non-English papers, 2) animal or cell studies, 3) reviews, case reports, and editorial articles, 4) studies that just concentrated on lncRNA genetic problems like polymorphism or epigenetic changes, 5) duplicated data.
2.3. Data Extraction
Two authors (MHA and ZB) undertook the data extraction from all the included studies independently. Any discrepancies were resolved by consensus. For each study, the first author's name and the year of publication, country. UC type, tumor size, sample size, lymph node metastasis (LNM), distance metastasis (DM), TNM stage, laboratory method in lncRNA expression, and HR with its 95% CI were recorded. The quality of data was evaluated by two authors according to the Newcastle-Ottawa Scale (NOS).
2.4. Statistical Analysis
The pooled HR and 95% CI were considered to evaluate the effect of lncRNAs expression on UCs (overall survival of multivariate analysis). An observed HR >1 suggested a worse prognosis. Also, we used an odd ratio (OR) with 95% CI to estimate the association between lncRNAs and clinicopathological parameters. In some publications, we extracted HRs from Kaplan Meier curves according to the methods reported by Parmar [11]. Cochran's Q test and I-squared statistics were performed to determine Heterogeneity between studies. The random-effect model was selected if the Heterogeneity was significant (I2>50% and P value<0.05); otherwise, the fixed-effect model was used. Subgroup and sensitivity analyses were done to investigate the source of heterogeneity. The Egger's test was performed to determine publication bias. P-value less than 0.05 is defined as statistical significance. Comprehensive Meta-Analysis software (CMA) was used for all analyses.
3. RESULTS
3.1. Literature Search
The detailed process of screening and the flow diagram of study selection is shown in Fig. (1). We explored 185 studies in the PubMed, Web of sciences, Scopus, and Google scholar databases. Initially, 42 duplicated articles were excluded in the initial search. After screening the titles and abstracts, 97 papers were excluded as basic research and insufficient data; finally, 46 eligible articles with 7597 patients were included in the current meta-analysis.

3.2. Study Characteristics
The main features and data of the selected articles are summarized in Table 1. These studies investigated a total of 4597 cases among these 46 studies, and all were from china. A total of 39 lncRNAs were explored in these 46 included studies. Three different types of UCs including bladder cancer (BC) [9, 12-27], PC [28-42], and renal cell carcinoma (RCC) [10, 43-53] were evaluated in this study. LncRNAs expression levels were measured in tumor tissues by quantitative Real-Time Polymerase Chain Reaction (qRT-PCR). The HR values were directly reported from thirty studies while, for fourteen studies, the HRs were considered through data reading from Kaplan-Meier survival curves. In addition, twenty-nine articles provided data regarding the association between lncRNAs expression and LNM, sixteen articles described DM, twenty sis papers reported TNM stage, and twenty-six studies reported tumor size.
| Study Year |
Age (High/ Low) |
Country | Tumor Type |
Tumor Size (High/ Low) |
Sample Size |
Male (High/Low) Female (High/Low) |
TNM Stage of Tumor (High/Low) |
Expression | Cut off Value |
Survival Analysis |
HR (CI) | Method | LncRNA name |
Sample Type |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High(n) | Low(n) | ||||||||||||||||||
| Total | LNM | DM | Total | LNM | DM | ||||||||||||||
| Shen 2020 | ≥65 5/36 <65 3/20 |
China | BC | ≥3 cm 2/30 <3 cm 6/26 |
64 | 8/49 0/7 |
NR | 8 | 0 | - | 56 | 26 | - | NR | OS | 0.203 (0.05-0.86) | RT-qPCR | Lnc-8996 | Tissue |
| Shen 2020 | ≥65 5/36 <65 6/17 |
China | BC | ≥3 cm 5/36 <3 cm 6/17 |
64 | 9/48 2/5 |
NR | 11 | 0 | - | 53 | 26 | - | NR | OS | 0.15 (0.03-0.73) | RT-qPCR | Lnc-4265 | Tissue |
| Li 2020 | ≥60 36/31 <60 14/17 |
China | BC | NR | 98 | 34/28 16/20 |
NR | 50 | 47 | - | 40 | 24 | - | NR | OS | 2.381 (1.821–7.012) | RT-qPCR | Lnc- PVT1 | Tissue |
| Zhang 2020 | >50 26/29 ≤50 27/24 |
China | BC | >2cm 39/25 ≤2cm 14/28 |
106 | 44/39 9/14 |
I/II 17/31 III/IV 36/22 |
53 | 31 | 38 | 53 | 19 | 27 | NR | OS | 2.27 (1.26-4.06) | RT-qPCR | Lnc-PCAT6 | Tissue |
| Cai 2020 | NR | China | RCC | ≥4cm 20/6 <4cm 4/17 |
47 | 17/14 7/9 |
I/II 9/11 III/IV 15/12 |
24 | - | 19 | 23 | - | 6 | NR | OS | 1.503 (0.314-7.196) | RT-qPCR | Lnc-PCGEM1 | Tissue |
| Bai 2020 | ≥65 39/33 <65 25/33 |
China | PC | NR | 130 | NR | I/II 23/40 III/IV 41/26 |
64 | 30 | 21 | 66 | 15 | 7 | NR | OS | 2.111 (1.735–10.295) | RT-qPCR | Lnc- NEAT1 | Tissue |
| Sun 2020 | ≥60 46/57 <60 95/85 |
China | RCC | NR | 283 | 98/96 43/46 |
NR | 114 | - | - | 103 | - | - | OS | 0.982 (0.444-2.171) | RT-qPCR | Lnc- APOC1P1 | Tissue | |
| Xia 2020 | ≥60 49/39 <60 17/22 |
China | PC | NR | 127 | NR | I/II 29/42 III/IV 37/19 |
66 | 24 | - | 61 | 9 | - | NR | OS | 2.839 (1.921–8.382) | RT-qPCR | Lnc- SNHG7 | Tissue |
| Duan 2019 | ≥60 21/14 <60 20/27 |
China | RCC | >7cm 10/4 ≤7cm 31/37 |
82 | 23/36 5/18 |
I/II 30/39 III/IV 11/9 |
41 | - | - | 41 | - | - | NR | OS | 3.23 (0.6-3.2) | RT-qPCR | Lnc-LINC-PINT | Tissue |
| Qian 2019 | ≥55 44/39 <55 48/51 |
China | RCC | ≥ 4 cm 32/40 <4 cm 60/50 |
182 | 52/58 40/32 |
I/II 58/48 III/IV 34/42 |
92 | 18 | 19 | 90 | 34 | 31 | NR | OS | 2.897 (1.275-4.387) | RT-qPCR | Lnc- PGM5-AS1 | Tissue |
| Shan 2019 | ≥50 28/23 <20/25 |
China | BC | ≥3cm 23/24 <3cm 25/24 |
96 | 34/28 14/20 |
I/II 12/30 III/IV 36/18 |
48 | - | 7 | 48 | - | 1 | NR | OS | 2.102 (1.145‐3.859) | RT-qPCR | FAM83H‐AS1 | Tissue |
| Zhu 2019 | >60 24/27 ≤60 30/28 |
China | PC | NR | 109 | NR | I/II 34/42 III/IV 20/13 |
54 | 15 | - | 55 | 5 | - | NR | OS | 2.985 (1.238-4.015) | RT-qPCR | Lnc- FEZF1- AS1 | Tissue |
| Li 2019 | ≥65 22/22 <60 31/30 |
China | PC | ≥2.5cm 26/20 <2.5cm 27/32 |
105 | NR | I/II 28/31 III/IV 25/21 |
53 | 17 | 25 | 52 | 11 | 13 | OS | 2.984 (1.263-4.198) | RT-qPCR | Lnc-LINC00662 | Tissue | |
| Xu 2019 | NR | China | PC | NR | 70 | NR | NR | 35 | - | - | 35 | - | - | NR | OS | 0.47 (0.19-1.152) | RT-qPCR | Lnc-TUG1 | Tissue |
| Wang 2019 | NR | China | BC | NR | 112 | NR | NR | 56 | - | - | 56 | - | - | NR | OS | 3.45(1.93-6.16) | RT-qPCR | Lnc- OIP5‐AS1 | Tissue |
| Xu 2019 | NR | China | BC | NR | 82 | NR | NR | 48 | - | - | 34 | - | - | NR | OS | 0.45 (0.19-1.02) | RT-qPCR | Lnc-NOTCH | Tissue |
| Li 2018 | ≥ 65 55/56 <65 13/18 |
China | PC | NR | 141 | NR | NR | 67 | 21 | - | 74 | 55 | - | NR | OS | 5.047 (4.863-5.488) | RT-qPCR | Lnc- BDNF-AS | Tissue |
| Huang 2018 | >60 25/45 ≤60 68/83 |
China | PC | NR | 54 | NR | NR | 93 | 45 | - | 128 | 31 | - | NR | OS | 3.302(1.380-7.89) | Lnc- TMPO-AS1 | Tissue | |
| Zheng 2018 | ≥65 32/19 <65 16/8 |
China | PC | ≥2 cm 32/10 <2 cm 16/17 |
75 | NR | I/II 34/20 III/IV 14/7 |
48 | 5 | 32 | 27 | 6 | 7 | NR | OS | 2.134 (1.214–3.753) | RT-qPCR | Lnc-NAP1L6 | Tissue |
| Zheng 2018 | ≥65 16/18 <65 11/8 |
China | PC | ≥2.5 cm 15/12 <2.5cm 12/1 |
53 | NR | I/II 6/18 III/IV 21/8 |
27 | 10 | - | 26 | 3 | - | NR | OS | 2.63(1.16-5.96) | RT-qPCR | Lnc-HULC | Tissue |
| Ma 2018 | ≥60 19/22 <60 17/9 |
China | BC | NR | 67 | 14/19 22/12 |
I/II 11/21 III/IV 25/10 |
36 | 27 | 21 | 31 | 11 | 8 | NR | OS | 3.68(1.17-11.54) | RT- qPCR | Lnc-SNHG5 | Tissue |
| Li 2018 | ≥60 28/37 <60 9/16 |
China | BC | ≥3cm 13/14 <3cm 24/39 |
90 | 31/37 6/16 |
I/II 14/33 III/IV 23/20 |
37 | 7 | - | 53 | 3 | - | NR | OS | 1.30 (1.01-1.68) | RT-qPCR | Lnc-NORAD | Tissue |
| Cao 2018 | ≥60 13/9 <60 12/12 |
China | BC | NR | 46 | 17/15 8/6 |
I/II 6/15 III/IV 19/6 |
25 | 17 | 20 | 21 | 2 | 8 | NR | OS | 3.21 (1.28-8.06) | RT-qPCR | Lnc- SNHG16 | Tissue |
| Su 2018 | ≥55 16/16 <55 14/15 |
China | RCC | NR | 61 | 21/25 9/6 |
I/II 25/15 III/IV 5/16 |
30 | 0 | 0 | 31 | 3 | 4 | NR | OS | 5.378 (2.084-13.88) | RT-qPCR | Lnc-ENSG00000241684 | Tissue |
| Liu 2018 | >60 22/23 ≤11/4 |
China | BC | >3cm 29/19 ≤3cm 6/6 |
60 | 32/21 3/4 |
NR | 35 | - | - | 25 | - | - | NR | OS | 3.729 (1.127-12.346) | RT-qPCR | Lnc-n346372 | Tissue |
| Zhang 2017 | ≥60 29/20 <60 16/7 |
China | PC | ≥2.5cm 18/11 <2.5cm 27/16 |
72 | NR | NR | 45 | 9 | - | 27 | 4 | - | NR | OS | 4.871 (2.604–9.110) | RT-qPCR | Lnc-LOC44004 | Tissue |
| Yang 2017 | ≥65 16/17 <65 13/12 |
China | BC | NR | 58 | 26/25 3/4 |
I/II 12/21 III/IV 17/8 |
29 | 7 | - | 29 | 3 | - | NR | OS | 0.379 (0.152-0.947) | RT-qPCR | Lnc-ASAP1-IT1 | Tissue |
| Yang 2017 | >60 42/21 ≤60 47/42 |
China | PC | NR | 152 | NR | I/II 45/31 III/IV 44/32 |
89 | 43 | - | 63 | 15 | - | NR | OS | 2.14 (1.29-3.55) | RT-qPCR | Lnc-PVT1 | Tissue |
| Wu 2017 | ≥65 44/12 <65 10/4 |
China | PC | NR | 73 | NR | NR | 54 | 33 | - | 16 | 5 | - | NR | OS | 4.003 (1.038–15.67) | RT-qPCR | Lnc-LINC01296 | Tissue |
| Shi 2017 | >60 11/9 <60 7/9 |
China | RCC | >3cm 17/5 <3cm 5/9 |
36 | 12/9 7/8 |
I/II 6/13 III/IV 13/4 |
19 | - | - | 17 | - | - | NR | OS | 1.93 (0.73-5.17) | qRT-PCR | Lnc-ROR | Tissue |
| Ning 2017 | >55 19/27 ≤55 18/12 |
China | RCC | >5cm 29/18 ≤5cm 8/21 |
76 | 15/19 22/20 |
NR | 37 | 18 | 28 | 39 | 6 | 27 | NR | OS | 2.13 (1.102-4.15) | qRT-PCR | Lnc-NEAT1 | Tissue |
| Li 2017 | ≥60 24/22 <60 40/34 |
China | BC | ≥3cm 29/28 <3cm 35/28 |
120 | 28/26 36/30 |
I/II 20/32 III/IV 44/24 |
64 | 21 | - | 56 | 7 | - | NR | OS | 2.056 (1.236-3.879) | qRT-PCR | Lnc- MALAT1 | Tissue |
| Du 2017 | ≥60 16/13 <60 29/21 |
China | BC | ≥3cm 17/18 <3cm 28/16 |
79 | 25/21 20/13 |
I/II 20/7 III/IV 25/27 |
45 | 9 | - | 34 | 13 | - | NR | OS | 0.52 (0.23-1.18) | qRT-PCR | Lnc- NBAT1 | Tissue |
| Zhao 2017 | ≥70 23/20 <70 20/22 |
China | PC | NR | 85 | NR | NR | 43 | 10 | 6 | 42 | 12 | 8 | NR | OS | 0.15 (0.028-0.791) | qRT-PCR | Lnc- FALEC | Tissue |
| Xiao 2017 | >60 124/106 ≤60 100/118 |
China | RCC | NR | 448 | 152/135 72/89 |
I/II 112/148 III/IV 112/76 |
224 | - | 46 | 224 | - | 25 | OS | 1.124 (1.043-1.211) | qRT-PCR | Lnc-Lucat1 | Tissue | |
| Zhang 2016 | ≥65 19/15 <65 23/18 |
China | BC | ≥2cm 29/16 <2cm 13/17 |
75 | 37/27 5/6 |
I/II 42/33 |
42 | - | - | 33 | - | - | NR | OS | 2.362 (1.504-4.83) | qRT-PCR | Lnc-UNMIBC | Tissue |
| Bao 2017 | ≥60 21/30 <60 37/41 |
China | RCC | >7cm 22/10 ≤7cm 36/61 |
129 | 49/48 9/23 |
I/II 33/63 III/IV 25/8 |
58 | - | - | 71 | - | - | NR | OS | 4.445 (1.396–14.153) | qRT-PCR | Lnc-PVT1 | Tissue |
| Wang 2016 | ≥65 33/22 <65 17/9 |
China | PC | ≥2.5cm 18/16 <2.5cm 32/15 |
81 | NR | I/II 34/25 III/IV 16/6 |
50 | 7 | 27 | 31 | 5 | 6 | NR | OS | 2.116 (1.231–3.638) | qRT-PCR | Lnc- LOC400891 | Tissue |
| Wu 2016 | >60 15/19 ≤60 23/20 |
China | RCC | >4cm 24/25 ≤4cm 14/14 |
77 | 25/23 13/16 |
I/II 6/16 III/IV 32/23 |
38 | - | 8 | 39 | - | 12 | NR | OS | 2.577 (1.233-5.387) | qRT-PCR | Lnc- Linc00152 | Tissue |
| Zheng 2016 | ≥65 40/25 <60 19/12 |
China | PC | ≥2.5cm 22/20 <2.5cm 37/18 |
96 | NR | I/II 14/22 III/IV 46/14 |
59 | 10 | 32 | 37 | 5 | 8 | NR | OS | 2.292 (1.370 -3.528) | qRT-PCR | Lnc- CCAT2 | Tissue |
| Xue 2015 | ≥65 27/18 <65 22/31 |
China | RCC | ≥4 cm 18/19 <4cm 31/30 |
98 | 26/28 23/21 |
I/II 38/25 III/IV 11/24 |
49 | 3 | - | 49 | 11 | - | NR | OS | 3.701(1.261-9.784) | qRT-PCR | Lnc-NBAT-1 | Tissue |
| Chen 2015 | >60 31/40 ≤60 21/17 |
China | BC | >3cm 31/34 ≤3cm 13/17 |
98 | 31/40 13/11 |
NR | 44 | - | - | 51 | - | - | NR | OS | 2.377 (1.007-5.610) | qRT-PCR | Lnc- n336928 | Tissue |
| Zhao 2015 | ≥ 59 14/12 <59 24/18 |
China | BC | ≥3 cm 17/14 <3cm 21/16 |
68 | 22/16 16/14 |
NR | 38 | 18 | - | 30 | 1 | - | NR | OS | 3.716 (2.084-6.719) | qRT-PCR | Lnc-SPRY4-IT1 | Tissue |
| Zhang 2014 | ≥ 65 25/20 <65 27/26 |
China | RCC | ≥ 4 cm 20/17 <4cm 32/29 |
98 | 29/25 23/21 |
I/II 26/37 III/IV 26/9 |
52 | 13 | 14 | 46 | 1 | 2 | NR | OS | 3.829 (2.274-8.178) | qRT-PCR | Lnc- SPRY4-IT1 | Tissue |
| Yan 2014 | ≥59 22/5 <59 68/15 |
China | BC | ≥3.5cm 28/7 <3.5cm 62/13 |
110 | 63/17 27/3 |
NR | 90 | - | - | 20 | - | - | NR | OS | 4.712 (2.894–8.714) | qRT-PCR | Lnc-HOTAIR | Tissue |
| Yao 2014 | >60 21/19 ≤60 11/13 |
China | RCC | ≥4cm 22/24 <4cm 10/8 |
64 | 18/20 14/12 |
I/II 30/24 III/IV 2/8 |
32 | - | - | 32 | - | - | NR | OS | 0.211(0.088-0.504) | qRT-PCR | Lnc- CADM1-AS1 | Tissue |
BC: Bladder Cancer, PC: Prostate Cancer, RCC: Renal Cell Carcinoma, DM: Distance Metastasis, LNM: Lymph Node Metastasis, OS: Overall Survival
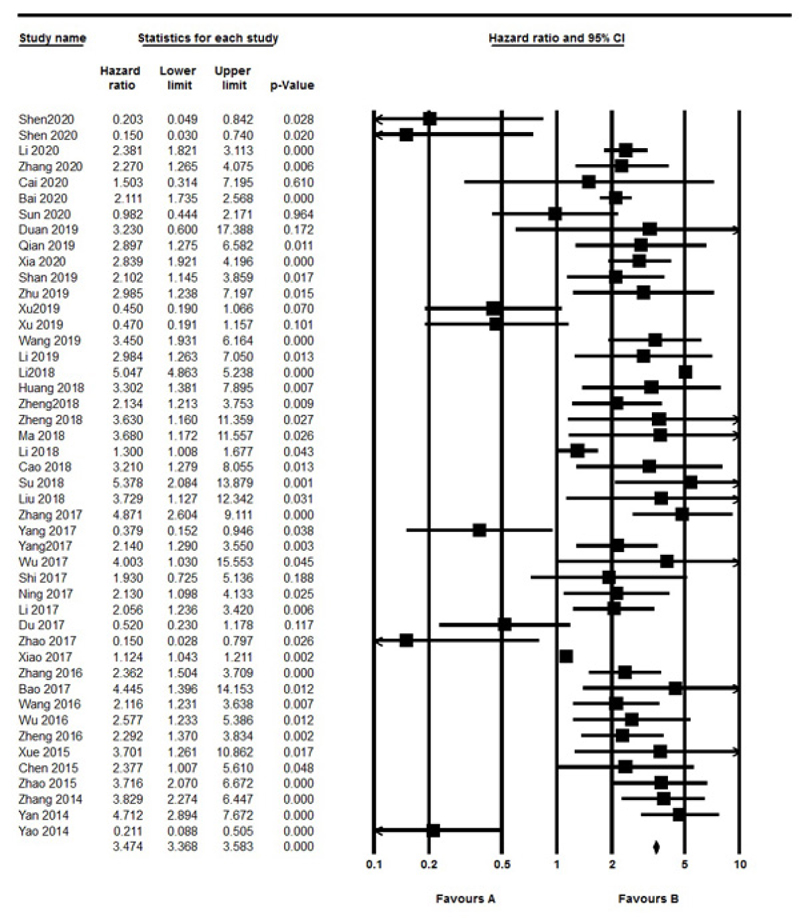
3.3. Association between lncRNAs Level and OS in UCs
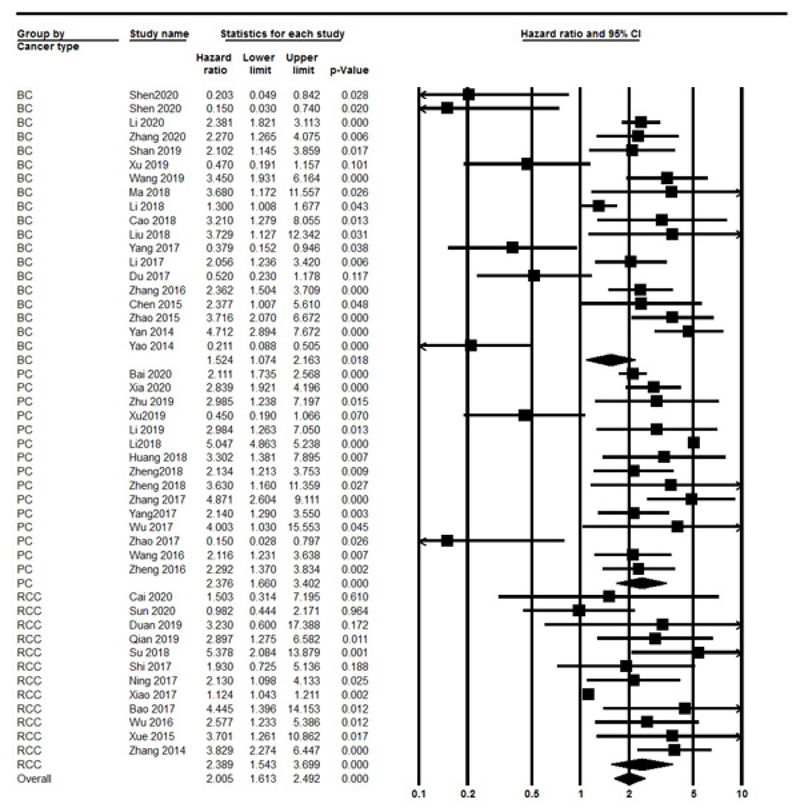
The OS was evaluated in 44 studies, respectively. Significant heterogeneity between included studies was found (I2=97.13%, P<0.001), therefore, the random effect model was performed to calculate the pooled HR and 95% CI. As a result, the pooled HR for all the studies showed a significant association between different lncRNAs expression and poor OS outcome (HR: 1.923, 95% CI: 1.448-2.554, P<0.001) (Fig. 2). Also, we divided included studies into up-regulated and down-regulated subgroups according to lncRNAs expression in UC. The results indicated a significant association with poor OS outcomes in both up-regulated (HR=2.546, 95% CI: 1.896-3.41, P<0.001, I2=97.3% P<0.001, Random model) and down-regulated (HR=0.33, 95% CI: 0.22-0.49, P<0.001, I2=0% P=0.4, Fix model) group. Also, we found a significant association between lncRNAs with sample size (≥100 vs. <100) (OR=1.893, 95% CI: 1.492-2.400, P<0.001, I2: 97.13% P<0.001, Random model). Furthermore stratified analysis according to urology cancer types revealed a significant association between the expression of lncRNAs with BC (HR=1.524, 95% CI: 1.074-2.163, P=0.018), PC (HR= 2.376, 95% CI: 1.66-3.4, P<0.001) and RCC (HR=2.389, 95% CI: 1.543-3.69, P<0.001) (Fig. 3).
3.4. LncRNAs Expression and Clinicopathological Characteristics
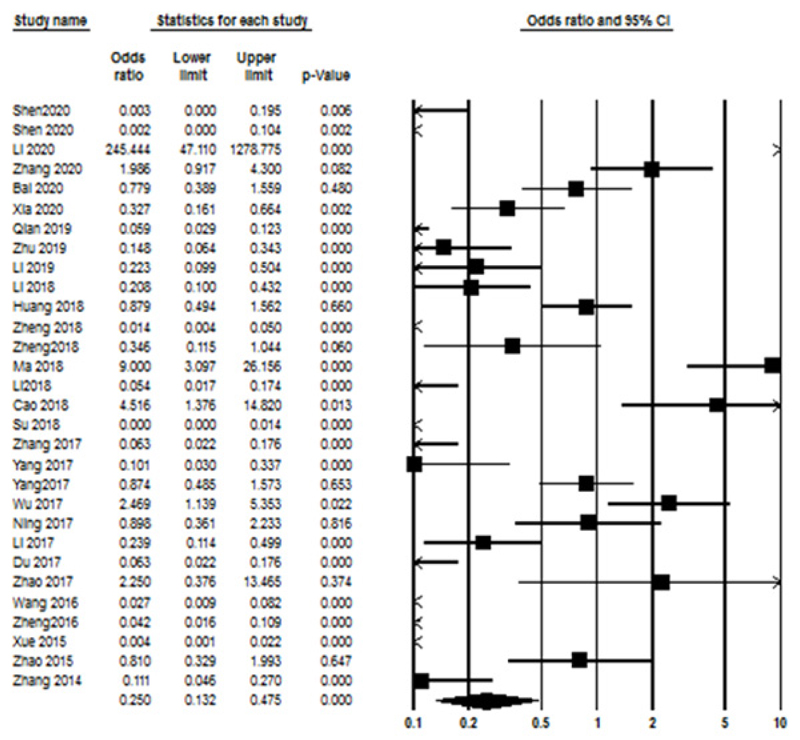
The association between lncRNAs expression level and the clinicopathological parameters was assessed by calculating the ORs, and the main results are summarized in Table 2. Accordingly, no significant correlation was seen for age (OR=1.301, 95% CI: 0.88-1.92, P=0.18, I2=89.23% P<0.001, Random model), tumor size (OR=1.159, 95% CI: 0.66-2.015, P=0.6, I2=90.307% P<0.001, Random model), TNM stage (OR=1.359, 95% CI: 0.8—2.309, P=0.25, I2=91.17% P<0.001, Random model), and DM (OR=0.507, 95% CI: 0.187-1.374, P=0.18, I2=95.35% P<0.001, Random model). Also, a significant relationship between lncRNAs expression and LNM (OR=0.25, 95% CI: 0.13-0.47, P<0.000, I2=92.140% P<0.001, Random model) (Fig. 4). The association between lncRNAs expression and clinicopathological characteristics are summarized in Table 2.
| Stratified Analysis |
No. of Studies |
No. of Patients |
Test of Association | Test of Heterogeneity | |||
|---|---|---|---|---|---|---|---|
| Pooled OR (95% CI) | p-value | I2 (%) | P-value | Model | |||
| Age (≥60 vs. <60) | 42 | 4286 | 1.301 (0.88-1.92) | 0.18 | 89.23% | <0.001 | R |
| Tumor size (large vs. small) |
28 | 2401 | 1.159 (0.66-2.015) | 0.6 | 90.307% | <0.001 | R |
| Tumor stage (III+IV vs. I+II) |
28 | 2909 | 1.359 (0.8-2.309) | 0.25 | 91.17% | <0.001 | R |
| DM | 17 | 1879 | 0.507 (0.187-1.374) | 0.18 | 95.35% | <0.001 | R |
| LNM | 30 | 2546 | 0.25 (0.13-0.47) | 0.000 | 92.140% | <0.001 | R |
3.5. Sensitivity Analysis and Publication Bias
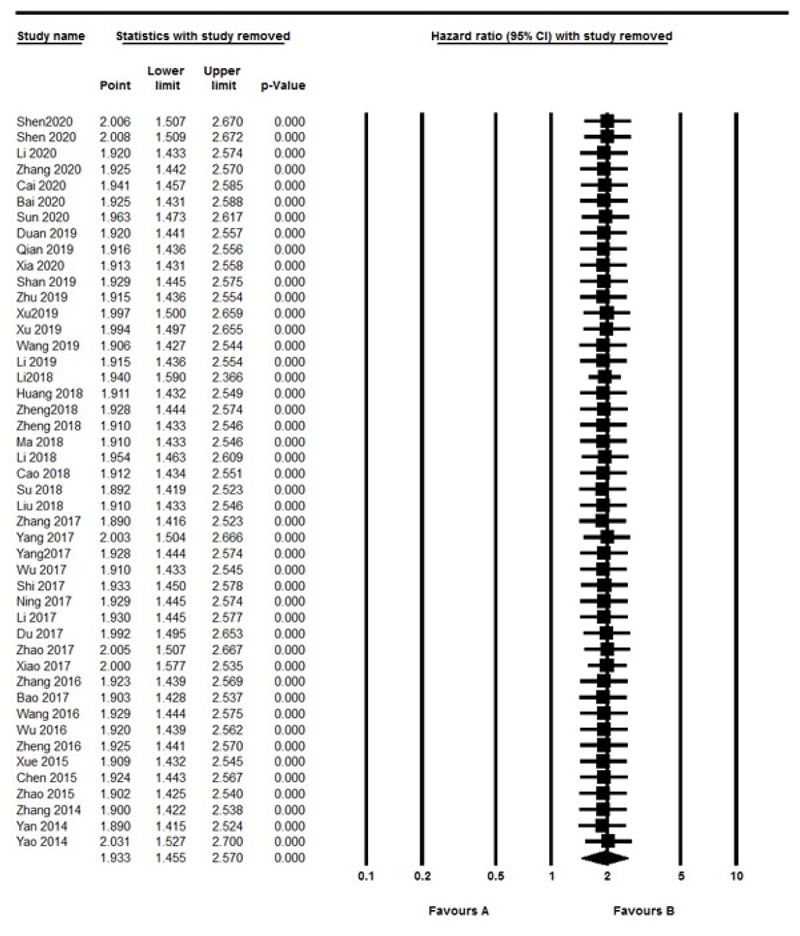
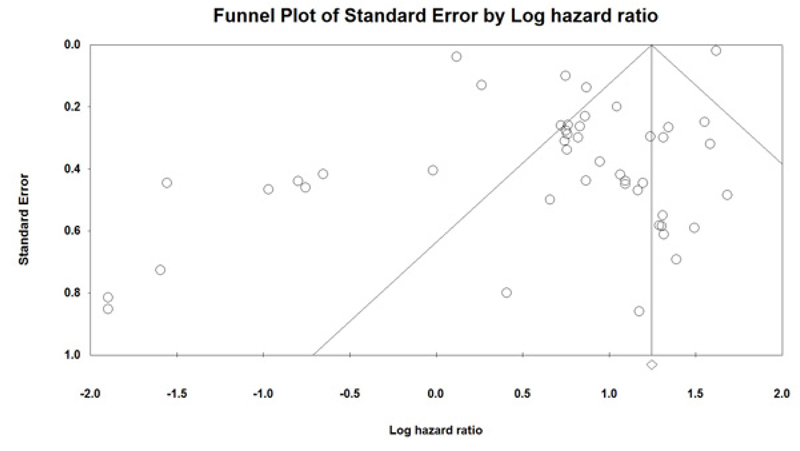
Sensitivity analysis was performed to assess the effect of each independent study on overall outcomes. After removing any single study, there was no significant change observed, so the results revealed the stability and robustness of the pooled results (Fig. 5). Moreover, the Funnel plot and Egger's tests were also used to evaluate the publication bias. In this Meta-analysis, the graphic assessment of the funnel plot and Egger's test indicated that there was significant publication bias through the included studies (P=0.01) (Fig. 6). When we removed six studies [12, 17, 24, 32, 40, 54], recalculated result showed no publication bias (P=0.1).
4. DISCUSSION
Although there have been substantial developments in the therapy of UCs in past decades, they are often detected at a late stage, because of the lack of effective biomarkers. Moreover, the evaluation of cancer prognosis is required for the selection of treatment type and understanding the disease process and survival rate [55]. UCs are associated with significant morbidity and mortality. The growing incidence of UCs in the western world indicates that these types of cancer will remain a significant health problem in the future. Therefore, the identification of new and effective biomarkers UCs is a crucial clinical need.
Recently, various studies focused on the clinical roles of lncRNAs, and many investigations showed that lncRNAs have the potential to be a molecular biomarker in UCs patients for predicting prognosis. About 15,000 lncRNAs have been found to exist in the genome of humans, and many of these are involved in carcinogenesis [4]. Many studies have demonstrated that lncRNAs can act as a transcriptional regulator playing roles in many cells processes such as cell growth and apoptosis-related to cancer. For instance, Cao et al. showed that lncRNA-SNHG16 promotes the proliferation of BC cells by affecting G1/S transition and reduce apoptosis. Also, they found that high-level expression of SNHG16 was associated with the overall survival of patients with BC [56]. Li et al. indicated that lncRNA LINC00662 by targeting miR-34a promotes the PC tumor genesis. They found a positive association between LINC00662 with clinical features such as distance metastasis and poor survival in patients with PC [31]. In addition, some lncRNAs have the role of tumor suppressors. Recently recognized lncRNA NBAT1 was able to control neuroblastoma and breast cancer progression through regulating cell proliferation [57, 58]. Du et al. showed reduced expression of lncRNA NBAT1 was related to promoting cell proliferation and epithelial mesangial transition and poor prognosis in patients with BC [24]. Our result showed that the down-regulation of 7 lncRNAs includes TUG1, NOTCH1, NBAT1, CADM1-ASP, FALEC, ENST00000598996 and ENST00000524265, were associated with poor OS in patients with UC.
This meta-analysis has summarized the current studies on the relationship between abnormal lncRNAs expression and the prognosis of patients with UC. Our result revealed a significant association between lncRNAs expression and poor prognosis in patients with UCs in both up-regulated and down-regulated groups. Next, we analyzed the association between lncRNAs level and the clinical features in patients with UC. LncRNAs expression was closely related to LNM and sample size but not associated with AGE, tumor size, TNM stage, and DM. In general, the pooled data showed that lncRNAs expression may represent a significant prognostic factor for survival outcomes in urological cancers.
There are some limitations to our meta-analysis. First, we estimated the HR and 95% CI from Kaplan-Meier curves by using DigitizeIT software and the methods reported by Parmar, which may also have affected the accuracy of our results. Second, all included studies in our meta-analysis were from China, our data might not be globally appropriate, third, the data were of various lncRNAs, which may have promoted the heterogeneity between the studies and conflict with the result.
CONCLUSION
This meta-analysis explored the relationship between the expression of lncRNAs and prognosis in UCS. Abnormal lncRNAs expression is linked to the poor prognosis of UCs patients and can therefore be a probable marker for UC. Although our knowledge about the biological effects of lncRNAs is limited, lncRNAs are probable have the potential to be biomarkers for UC. Finally, we need more studies with larger samples to confirm the conclusion of our meta-analysis in the future.
LIST OF ABREVIATIONS
| BC | = Bladder Cancer |
| CI | = Confidence Interval |
| DM | = Distance Metastasis |
| HRs | = Hazard Ratios |
| lncRNAs | = Long Non-coding RNAs |
| LNM | = Lymph Node Metastasis |
| NOS | = Newcastle-Ottawa Scale |
| ORs | = Odds Ratios |
| OS | = Overall Survival |
| PC | = Prostate Cancer |
| qRT-PCR | = Quantitative Real-time Polymerase Chain Reaction |
| UC | = Urological Cancer |
| RCC | = Renal Cell Carcinoma |
AUTHORS' CONTRIBUTIONS
MH.A and ZB researched literature and interpretation of data, MHA, final approval of the version to be published.
CONSENT FOR PUBLICATION
Not applicable.
STANDARDS OF REPORTING
PRISMA guidelines were followed in this study.
FUNDING
This study was supported by grants awarded by the Shahrekord University of Medical Sciences (Grant No. 5646).
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
ACKNOWLEDGMENTS
We thank the Shahrekord University of Medical Science to provide conditions for access to databases.


