All published articles of this journal are available on ScienceDirect.
COL1A1 Gene Expression in Hepatitis B Virus (HBV) Related Hepatocellular Carcinoma (HCC) Egyptian's Patients
Abstract
Introduction:
Collagens are the most abundant proteins in the human body, accounting for one-third of total proteins. Over the last few years, accumulated evidence have indicated that some collagens are differentially expressed in cancer. The aim of the study was to assess COL1A1 gene expression as a novel marker for the progression of hepatitis B cirrhosis into hepatocellular carcinoma.
Methods:
This cohort study included 348 subjects and was conducted between May 2018 and June 2019. Subjects were divided into 4 groups: group1 included HBV positive hepatocellular carcinoma patients “HCC” (n= 87), group II included HBV positive patients with liver cirrhosis “LC” (n = 87), group III included chronic hepatitis B patients with neither HCC nor cirrhosis “ C-HBV” (n = 87) and group IV consisted of healthy volunteers as controls (n = 87). Fasting venous blood samples (10 ml) were collected from each participant in this study and were used for assessment of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, albumin and alfa-fetoprotein (AFP). Another portion of blood was collected in 2 vacutainer tubes containing EDTA, one for Complete blood count and the other for gene expression of COL1A1.
Results:
The gene expression of collagen was 6.9 ± 8.8 in group 1 (HBV positive hepatocellular carcinoma patients) and this was a significant increase in comparison with the other groups. In group 2 (HBV positive patients with liver cirrhosis), the gene expression (collagen) was 3.7±1.5 and it was significantly increased when compared with group 4 (healthy volunteers).
Conclusion:
COL1A1 gene expression can be used as an indicator of the progression of hepatitis B cirrhosis into hepatocellular carcinoma.
1. INTRODUCTION
Hepatocellular Carcinoma (HCC) is one of the causes of death all over the world [1]. It is the third lethal type of cancer, causing around 600,000 deaths per year [2]. It represents the sixth most common cancer worldwide, and in Egypt, it represents the fourth [3]. It is a sequela of diverse causes of liver damage as chronic hepatitis C and B infection, chronic use of alcohol as well as nonalcoholic steatohepatitis [4] (EASL guidelines, 2018).
Chronic Hepatitis C virus (HCV) infection represents 75% to 85% of the HCV-infected patients, and nearly 20% of them will later develop complications like cirrhosis or HCC after 20 years of infection [5]. Although liver resection and transplantation are the most common treatments for HCC, high rates of recurrence and metastasis lead to poor prognosis. Therefore, early diagnosis and treatment of HCC are essential [6]. Hepatitis B is also a life-threatening liver infection due to the aggressive transformation to liver cirrhosis and HCC. The WHO previously reported Egypt an area of endemicity of intermediate degree (2%-7%) regarding HBV [7].
Collagens are the most abundant proteins in the human body, accounting for one-third of total proteins. In humans, there are at least 28 different types of collagen proteins encoded by 44 collagen genes [8]. They are essential in the extracellular matrix (ECM), which is the major component in the tumor microenvironment and can regulate tumor cell behaviors [9, 10]. Collagen proteins are produced and secreted by fibroblasts [11], osteoblasts [12], hematopoietic cells [13], and also can be biosynthesized and regulated by endothelial cells [14, 15] and cancer cells [16]. Over the last few years, accumulated evidence-have indicated that some collagens are differentially expressed in cancer. Collagen type I contributes to pancreatic, lung, bladder, liver and breast cancer progression [17-19], as it enhances the tumor cell proliferation and the epithelial-mesenchymal transition of HCC [20].
Collagen type I alpha 1 (COL1A1) gene encodes the pro-alpha 1 chains of type I collagen whose triple helix comprises two alpha 1 chains and one alpha 2 chain [21]. Type I collagen is a fibril-forming collagen, and the 3D type I collagen-rich culture is a common method to investigate the proliferation and/or metastasis of cancers [22]. Previous studies have suggested that COL1A1 is a candidate survival-related factor in hepatocellular carcinoma [23]. Collagen is also considered an important substance seen in liver fibrosis, especially collagen type IV, which is available as a marker of hepatitis C fibrosis. Collagen types I, II, and III have also been reported to be associated with the liver fibrosis stage of chronic HCV [23].
COL1A1 promotes Slug-dependent epithelial-to-mesenchymal transition, thereby enhancing the invasion and metastasis of HCC [24]. Recently, abnormal expression levels of COL1A1 and COL1A2 have been reported in several types of cancer [25]. COL1A1 and COL1A2 mRNA expression levels are upregulated in colorectal cancer and \\medulloblastoma. COL1A1 and COL1A2 were deferentially expressed in gastric cancer and predicted poor clinical outcomes in gastric cancer patients [26, 27].
2. MATERIALS AND METHODS
2.1. The Setting, Study Design
This cohort study included 348 subjects and was conducted between May 2018 and June 2019 in Tanta University Hospital and the National hepatology and tropical medicine research institute. The study was approved by the institutional ethical committee. Subjects were divided into 4 groups: group 1 included HBV positive hepatocellular carcinoma patients “HCC” (n= 87), group II included HBV positive patients with liver cirrhosis “LC” (n = 87), group III included chronic hepatitis B patients with neither HCC nor cirrhosis “C-HBV” (n = 87) and group IV of healthy volunteers as controls (n = 87).
For all subjects, complete medical history and physical examination were done, and body mass index (BMI) was calculated. All patients were positive for HBV, confirmed by hepatitis B surface antigen (HBsAg) and HBV PCR (Qiagen, MX 3000 applied biosystem). All subjects were confirmed as HCV negative by screening for Anti-HCV using a commercially available immunoassay (HCV Murex AB, Diasorin, Saluggia, Vercelli, Italy).
For HCC patients and LC patients, the liver status was estimated by Model for End-Stage Liver Disease (MELD) score and Updated MELD (U MELD) score. HCC patients were staged according to Okuda staging, Cancer of the Liver Italian Program (CLIP) staging and Vienna survival model for HCC (VISUM-HCC).
Study exclusion criteria were: previous cancer, current cancer apart from HCC cancer, history of alcohol abuse, renal insufficiency, proteinuria, suspected infections, clinically overt diabetes mellitus, thyroid dysfunction, or any other endocrine disorder, and hormone or thyroid regulatory medication intake. Informed consent was obtained from all participating subjects before the study. All participants gave their written informed consent prior to inclusion.
2.2. Blood Sampling and Biochemical Assays
Fasting venous blood samples (10 ml) were collected from each participant in this study. A portion of blood was left to clot and then centrifuged at 3500 g for 5 min to separate the serum used for assessment of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, albumin and alfa-fetoprotein (AFP). Another portion of blood was collected in 2 vacutainer tubes containing EDTA, one for Complete blood count and the other for gene expression of COL1A1.
2.2.1. RNA Extraction
Peripheral Blood Mononuclear Cells (PBMCs) were separated using Ficoll density centrifugation and sedimentation. RNA was separated from PBMCs using QIAamp viral RNA extraction kit (QIAGEN GmbH, Hilden, Germany).
2.2.2. Quantification of COL1A1
Gene expression of COL1A1 was performed using TaqMan Expression (Applied Biosystems Inc, Foster City, CA, USA). β-actin was the reference gene for each sample. Fractional threshold cycles (CT) expressed the initial concentration of the target sequence. Relative mRNA quantification was calculated using the arithmetic formula 2 –ΔC T, where ΔCT is the difference between the CT of a given target cDNA and an endogenous reference cDNA. Thus, this value yielded the amount of the target normalized to an endogenous reference [28].
2.3. Statistical Analysis
The collected data were tabulated and analyzed using SPSS version 20 software (SPSS Inc, Chicago, ILL Company). Categorical data were presented as numbers and percentages, while quantitative data were expressed as mean ±standard deviation. Chi-square (X2) test, Fisher's exact test, ANOVA, and Pearson’s correlation coefficient (r) were used as tests of significance. Significant ANOVA was followed by the Bonferroni test to detect significant pairs. ROC curve was used to determine the cutoff value of collagen with optimum sensitivity and specificity in early diagnosis of HCC. The accepted level of significance in this work was stated at 0.05 (P <0.05 was considered significant) [29].
3. RESULTS
The clinical characteristics of the included subjects are shown in Table 1, 64.4% (n = 56) were male. Smoking-induced liver disease was the most frequent underlying cause of HCC in 59.8% (n = 52).
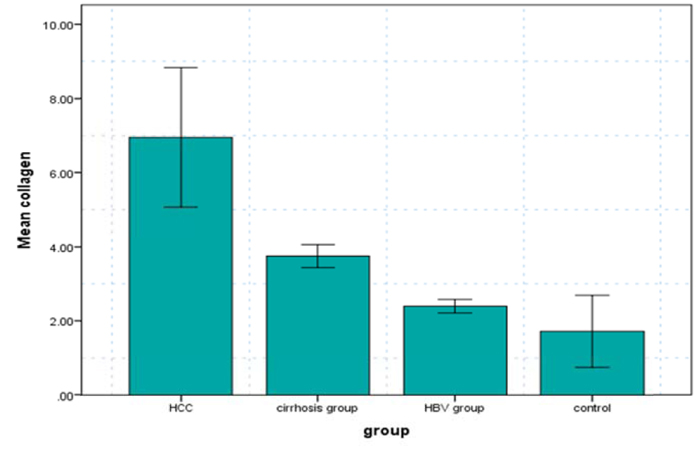
Serum levels of collagen were measured in each of the 4 groups. The comparison showed marked differences in collagen-related measures (Table 2). It was found to be significantly increased in HBV patients (2.4 ± 1.8 ng/ml), cirrhotic patients (3.7±1.5 ng/ml) and HCC patients (6.9±8.8ng/ml) in comparison to healthy subjects (1.7±4.5 ng/ml).
| I- HCC (87) | II- Cirrhosis Group (87) | III-HBV Group (87) | IV-Control (87) | P Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | No | % | |||
| Sex | Female | 31 | 35.6 | 46 | 52.9 | 29 | 33.3 | 33 | 37.9 | 8.5(0.04*) |
| Male | 56 | 64.4 | 41 | 47.1 | 58 | 66.7 | 54 | 62.1 | ||
| HBVsAg | Negative | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 100 | 348(.000*) |
| Positive | 87 | 100 | 87 | 100 | 87 | 100 | 0 | 0 | ||
| HCV ab | Negative | 87 | 100 | 87 | 100 | 87 | 100 | 87 | 100.0 | - |
| Positive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Alcohol | No | 86 | 98.9 | 78 | 89.7 | 76 | 87.4 | 87 | 100.0 | 18.8(.000*) |
| Yes | 1 | 1.1 | 9 | 10.3 | 11 | 12.6 | 0 | 0 | ||
| Smoking | No | 35 | 40.2 | 81 | 93.1 | 48 | 55.2 | 87 | 100 | 109(.000*) |
| Yes | 52 | 59.8 | 6 | 6.9 | 39 | 44.8 | 0 | 0 | ||
| - | I-HCC (87) | II-cirrhosis Group (87) | III-HBV Group (87) | IV-Control (87) | P Value |
|---|---|---|---|---|---|
| Age | 61.17±9.4 | 55.8±10 | 38.3±8.5 | 43.2±15.5 | 78.4(.000*) |
| ALT | 63.08± 33.8# (III, IV) | 53.6±57.9# (III, IV) | 79.4±29.9 | 30.3±5.6# (I, II, III) | 27(.000*) |
| AST | 103.4±65# (III, IV) | 81.8±119.6# (IV) | 67.4±30.9#(I, IV) | 33.2±7.6 | 15.5(.000*) |
| Total bilirubin | 4.5±4.9# (III, IV) | 4.9±5.8# (III, IV) | 1.1±0.4 | 0.76±0.2 | 28.5(.000*) |
| INR | 1.5±.3034# (3,4) | 1.5±0.8# (3,4) | 1.1±0.3 | 0.98±0.07 | 29.7(.000*) |
| Albumin | 2.614± 0.5# (III, IV) | 2.5±0.8#(III, IV) | 3.6±0.6 | 3.9±0.2 | 130.6(.000*) |
| ALP | 170.6±117.5 | 156.3±110.828 | - | - | 0.8(>0.05) |
| AFP | 2846.9±1342.2# (II, IV) | 208.2±143.4 | - | 5.7±1.7 | 17.3(.000*) |
| Serum creatinine | 2.15±1.7# (II, III, IV) | 1.6±1.2# (II, III, IV) | 1.023±0.3713#(III) | 0.9±0.16 | 24.5(.000*) |
| Hb | 11.190± 1.7 | 10.6±2# (I, III) | 12±1.8 | - | 12.4(.000*) |
| WBCS | 1476± 232.6 | 7.6±5.5 | 8034.5 ±1382.5# (II, I) | - | 1333(.000*) |
| Platelets | 118.414±53.2647 | 137±70.3 | 294.8±75.3# (II, I) | - | 182.2(.000*) |
| Meld | 20.4±9 | 18.1±9.3 | - | - | 1.6(>0.05) |
| U meld | 4.140±1 | 3.9±1.3 | - | - | 1.3(>0.05) |
| PV | 1.2±0.4 | - | - | - | - |
| Temp | 37.3±0.5 | - | - | - | - |
| Pulse | 85.09± 6.2 | - | - | - | - |
| BMI | 26.06±2.2 | 30.7±2.2 | 30.3±4.7#(I,IV) | 22.5±3.6 | 117.9(.000*) |
| Okuda stage | 2.32±0.7 | - | - | - | - |
| Okuda score | 2.2±1.3 | - | - | - | - |
| Clip stage | 2.3±0.6 | - | - | - | - |
| Clip score | 2.9± 1.6 | - | - | - | - |
| Tokyo score | 5.2±1.6 | - | - | - | - |
| Visme HCC viena stage | 1.7±0.8 | - | - | - | - |
| SME HCC Viena score | 2.4±1.4 | - | - | - | - |
| Size | 4.74±2.548 | - | - | - | - |
| HBV DNA | 3551.3± 2023.1 | 3383.6±1448.8 | 1455078694.3±6525410985.2#(I,II) | - | 4.3(0.02*) |
| Gene expression (collagen) | 6.9 ±8.8# (II,IIII,IV) | 3.7±1.5# (IV) | 2.4±1.8 | 1.7±4.5 | 18.5(.000* |
The collagen levels were higher in HCC and LC patients as compared with the other 2 groups. Moreover, the increase of collagen levels was also statistically significant between both HCC and LC patients and CH-B patients, and between HCC and LC patients, as shown in Fig. (1).
The Receiver Operating Characteristic (ROC) curve was used to determine the cut-off value of collagen for predicting the development of HCC in HBV-positive patients with liver cirrhosis. The area under the ROC curve (AUC) was 0.529, indicating a fair predictive power for hepatic cancer (Fig. 2).

4. DISCUSSION
HCC is one of the sequelae of liver damage due to chronic hepatitis C and B infection [29-42]. COL1A1 is a candidate survival-related factor in hepatocellular carcinoma, which was proved to enhance the invasion and metastasis of the neoplasm [10].
In this study, there were 3 groups of HBV-positive patients compared to the 4th group of healthy controls. The gene expression of collagen was 6.9 ± 8.8 in group 1 (HBV positive hepatocellular carcinoma patients), and this was a significant increase in comparison with the other groups. In group 2 (HBV positive patients with liver cirrhosis), the gene expression (collagen) was 3.7±1.5, and it was significantly increased when compared with group 4 (healthy volunteers).
ROC curve was made to detect the optimal cut-off value for predicting the occurrence of hepatocellular carcinoma, and it was found that the COL1A1 gene expression of ≥ 5.6 has a fair predictive power, which can predict the presence of HCC with 61.5% sensitivity and 40% specificity and 95% Confidence Interval.
These results are concordant with Ma et al. as well as Lin et al., who found that COL1A1 is extremely expressed in Hepatocellular carcinoma and deliberates substantial survival disadvantage, and that upregulated COL1A1 expression at both mRNA and protein levels strongly correlates with Hepatocellular carcinoma evolution [24, 30]. Also, Hayashi et al. found that the COL1A1 was highly expressed in cases of liver cirrhosis as well as HCC compared to the healthy population [23].
On the other hand, Hayashi et al. found that there was a small group of HCC that has down-regulated mRNA expression mainly due to promoter methylation, and these cases were correlated with poor overall survival. In these cases, the single nucleotide polymorphism array displayed no chromosomal deletion in the locus of COL1A1. Importantly, the methylation value in the tumor tissue was higher than that of the adjacent liver tissue [23].
Moreover, due to the epigenetic modifications, there are different expressions of COL1A1 in different tumors. Frequent promoter methylation was found in renal cell carcinoma [31], and there was a decreased expression in ovarian serous carcinoma [32].
It is consistent with recent conclusions that showed COL1A1 was highly expressed in human breast cancer. Additionally, COL1A1 mRNA levels are significantly elevated in gastric cancer tissues when compared to normal ones, and lower expression levels of COL1A1 are powerfully connected with improved survival in patients with gastric cancer [33].
Moreover, like our results, Zhao et al. found that the frequency of COL1A1 polymorphism was higher in cirrhotic patients as compared to healthy people. This was accompanied by higher transcriptional activity and a high possibility of association with liver fibrogenesis. But they did not investigate the marker in HCC [34]. Sun et al. studied the COL1A1 gene polymorphism in HCC and reported an association with a reduced COL1A1 mRNA expression which was significantly correlated with capsule formation in patients with HCC [23, 34, 35]. However, in our patients, we studied the gene expression rather than the polymorphism in the whole cancer group rather than a capsulated group.
CONCLUSION
In conclusion, COL1A1 gene expression can be used as an indicator of the progression of hepatitis B cirrhosis into hepatocellular carcinoma, and further studies are required on a large sample size.
Another study reported the expression of COL1A1, COL1A2, COL3A1 in blood and could be applied as predictive biomarkers in lung adenocarcinoma [36], which indicates the important role of this gene in many diseases.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Research Ethics Committee of Tanta University Faculty of Medicine, Tanta, Egypt with approval number: 30431/11/18.
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures were followed in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
All participants gave their written informed consent prior to inclusion.
AVAILABILITY OF DATA AND MATERIALS
Datasets used and/or analysed during the current study are available from the corresponding author [S.A.] on reasonable request.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


