All published articles of this journal are available on ScienceDirect.
Talin-1 Gene Expression as a Tumor Marker in Hepatocellular Carcinoma Patients: A Pilot Study
Abstract
Background & Aims:
Hepatocellular Carcinoma (HCC) is the most common primary liver tumor. It is the second most common cancer in men and the sixth in women in Egypt. One of the proteins participating in the trans-endothelial migration is Talin-1. It also has a role in the formation and metastasis of different types of cancer. This study aimed to evaluate the diagnostic impact of Talin-1 gene expression in HCC Egyptian patients.
Methods:
Our study included forty HCC patients, thirty liver cirrhosis patients without HCC and thirty healthy subjects. For all groups, clinical and biochemical parameters were investigated. Tumor characteristics were assessed and tumor staging was done using Okuda, CLIP, VISUM and Tokyo staging systems. In addition, Serum Alpha-Fetoprotein (AFP) levels were assayed using Enzyme Immunoassay (EIA) and Talin-1 gene expression was assessed in the Peripheral Blood Mononuclear Cells (PBMCs) via quantitative real-time Polymerase Chain Reaction (PCR).
Results:
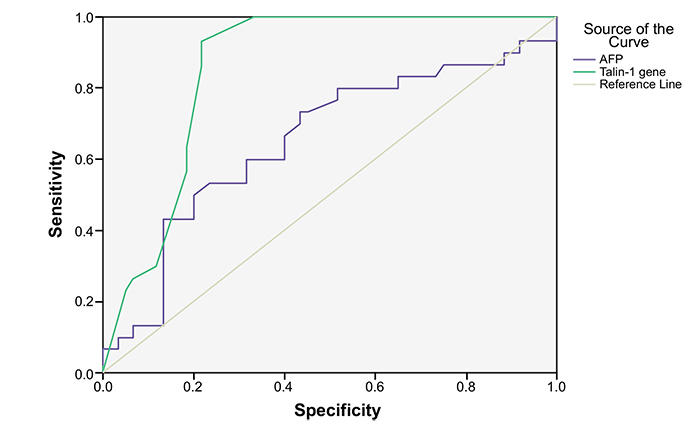
Talin-1 gene expression was significantly upregulated in HCC patients in comparison to cirrhotic and control subjects. The Receiver Operating Characteristic (ROC) analysis indicated that Talin-1 gene expression surpasses serum levels of AFP in the diagnosis of HCC. In particular, the cut off value of 9.5 (2-∆∆Ct) recorded an AUC of 85.7% with a sensitivity of 93.3% and specificity of 80%.
Conclusion:
Our data confirmed an évident diagnostic role of Talin-1 gene expression for HCC detection.
1. INTRODUCTION
Hepatocellular Carcinoma (HCC) is the fifth most common cancer in males, and the seventh between females all over the world. More than half a million cases are newly diagnosed every year [1]. In the developing countries, incidence and total mortality are representing 84% and 83% of the worldwide respectively [2].
In Egypt, the incidence of HCC has increased from 4% to 7.2% within the last ten years. This rise can be explained by the increase in risk factors, such as chronic Hepatitis C Virus (HCV) infection [3-10]. Curative treatment (e.g. hepatic resection, local ablation, or liver transplantation) is associated with better 5-year survival (70-80%). However, it is only available for less than 30% of patients, who are diagnosed in the early stages. In advanced cases, high rates of recurrence and metastasis make the prognosis worse. Therefore, the best strategy of surveillance programs is focused upon early diagnosis of HCC [11, 12].
Alpha-Fetoprotein (AFP) is a serological marker commonly used for the detection of HCC. However, its low specificity and sensitivity make it less reliable in early HCC diagnosis, prevention or therapy. Consequently, new and specific markers for HCC are critically needed [13-15].
Talin-1 is a large cytoskeletal dimeric adaptor protein270 kDa molecular weight. It activates integrins (family of cell adhesion molecules in cell-extracellular matrix junctions), then couples them to the actin cytoskeleton [15].
Once integrins activated, they initiate activation of Focal Adhesion Kinase (FAK), which regulates many processes relating to cancer development, including cell survival, proliferation, migration, invasion, and metastasis [16-18]. Nowadays, the effect of Talin1 in malignancies has drawn attention. Some studies detected that overexpression of Talin1 was associated with poor prognosis and a higher rate of portal vein invasion in HCCs [19-22].
This study aimed to evaluate the diagnostic impact of Talin-1 gene expression in HCC Egyptian patients in comparison to serum AFP levels in patients with hepatocellular carcinoma.
2. METHODS
The study included 70 patients with chronic liver disease, divided into two groups: Group (I) included forty patients with HCC; Group (II) included thirty patients with liver cirrhosis and without any evidence of HCC. Thirty healthy adults were recruited as control (Group III). Patients with cancers other than HCC or metastatic liver cancer were excluded. The current study was approved by the Ethics and Research Committee of Benha Faculty of Medicine, Benha University, Egypt. All patients had the procedure thoroughly explained prior to their enrollment in the study.
Hepatocellular carcinoma was suspected by the abdominal US and confirmed by triphasic CT scan with contrast [23]. The historical, clinical, and biochemical data of the patients were obtained, including age, gender, alcohol intakes, Hepatitis C Virus (HCV) and Hepatitis B Virus (HBV), liver function tests, and AFP levels. Assessment of liver cirrhosis was done using by modified Child-Pugh score [24], and MELD (Model for End-stage Liver Disease) score [25] and the updated MELD (uMELD) score [26]. Tumor characteristics were demonstrated by CT scan (including tumor size, number, site, halo sign, and neovascularization). Tumor staging was done using Okuda [27], CLIP (The Cancer of the Liver Italian Program) [28], VISUM (Vienna Survival Model for HCC) [29], and Tokyo [30] staging systems. Fig. (1) shows a flow chart of the study.
2.1. Blood Sampling and Biochemical Assays
All samples were collected during fasting. All assays were performed in duplicate according to the manufacturer’s instructions. Serum samples were aliquoted and stored at -80°C until assayed. Beckman CX4 chemistry analyzer (Diamond Diagnostics, NY, USA) was used to determine serum levels of Aspartate Aminotransferase (AST), Alanine Aminotransferase (ALT), total bilirubin, albumin, and creatinine. HCV Ab and HBS Ag were analyzed using Abbott, Axyam (Abbott Diagnostics, Santa Clara, California, USA). Serum AFP was assayed via an Enzyme Immunoassay (EIA) (Roche Mannheim, Germany). In addition, blood samples were collected in vacutainer tubes containing EDTA to separate lymphocyte cells for Talin-1 gene expression assay.
2.2. Quantitative Real-time PCR Assay
Talin-1 gene expression was analyzed in Peripheral Blood Mononuclear Cells (PBMCs), which were isolated from peripheral blood using Ficoll density centrifugation and sedimentation. Total RNA was extracted from PBMCs cells using QIA amp viral RNA extraction kit. The reverse transcription was carried out on the extracted RNA using the high capacity cDNA reverse transcription kit (Applied Biosystems, USA). Two 2 μg of total RNA were mixed with 0.5 μg of oligo (dT) 12-18 primer in a total volume of 12μL. The mixture was heated at 70°C for 10 min. A solution containing 50 mmol/L Tris HCl (pH 8.3), 75 mmol/L KCl, 3 mmol/L MgCl2, 10 mmol/L DTT, 0.5 mmol/L dNTPs, 0.5 μL RNase inhibitor, and 200 U Reverse Transcriptase was added, resulting in a total volume of 20.5 μL. All mixture was incubated at 42°C for 1 h. Quantification was performed using TaqMan® Gene Expression assay (Applied Biosystems Inc, Foster City, CA, USA) according to the manufacturer's instruction. The qRT-PCR was executed using the following primers: Talin-1 sense, 5’-TCTCCCAAA ATGCCAAGAAC-3’ and anti-sense, 5’-TGGCTATTGG GGTCAGAGAC-3’; Glyceraldehyde -3- phosphate dehydrogenase (GAPDH) sense, 5’-CCACTCC TCCACCTTTGAC-3’ and anti-sense, 5’-ACCCTGT TGCTGTAGCCA-3’ according to previously published sequences [31, 32]. The relative expression of Talin-1 was normalized to GAPDH as housekeeping gene and the Cycle Threshold (CT) was calculated. Relative levels of Talin-1 gene expression were calculated using the comparative threshold cycle method and expressed as 2-ΔΔCT [33]. Briefly, relative quantification of Talin-1 gene expression was normalized to the endogenous reference GAPDH and relative to a calibrator.
Statistical Analysis: Data were statistically analyzed using the statistical package (SPSS, version 20.0). Categorical data were presented using number and percentage and compared using the Chi-square test. Mean, and standard deviation were used for numerical measures. Unpaired student t-test and Mann-Whitney test were used to compare normal and not normal distributed numerical, respectively. Receiver Operating Characteristic curves (ROC) were constructed to assess the validity of the markers in predicting HCC by calculating the Area Under the Curve (AUC). Cut off values of different diagnostic markers were detected. Youden's J statistics was calculated for each cut-off point as follows [sensitivity + specificity -1], the selected cut-off point was the one that achieved the higher Youden's J value considering the higher sensitivity and specificity. Sensitivity, specificity, positive (PPV) and negative (NPV) predictive values were calculated. Probability < 0.05 was considered significant.

3. RESULTS
Demographic features as well as characteristics of HCC and cirrhosis patients are summarized in Table 1. Serum AST and ALT levels were significantly increased in HCC group versus the cirrhotic group, while the rest of biochemical parameters were statistically insignificant between all groups.
No statistically significant changes were shown among HCC and cirrhotic groups as regards the severity of the liver disease assessed by Child-Pughh classification; MELD score and uMELD score (Table 2).
The tumor-related characteristics of HCC patients are illustrated in (Table 3). HCC was single lesion in (57.5%) of patients, large in (40%), in the right lobe in (60%), halo sign in 95%, and Portal Vein Thrombosis (PVT) in (12.5%). Regarding Okuda and Tokyo staging systems, most HCC patients were relatively at advanced stages. Additionally, as regard the correlation of Talin-1 relative expression with Child score, MELD, uMELD, tumor number, tumor size, AFP, Okuda, CLIP, Tokyo, and VISUM staging systems, no statistically significant correlation was detected.
Talin-1 gene expression was significantly up-regulated in HCC patients in comparison to cirrhotic and control groups. Regarding AFP levels, both HCC and cirrhotic patients recorded significantly higher levels versus control group. Whereas, there was an insignificant change in AFP levels between HCC and cirrhotic patients (Table 4).
ROC curve analysis showed that the area under the curve of Talin-1 gene expression was higher than that of AFP (Fig. 2). The diagnostic validity and the optimal cut off values of Talin-1 gene expression and AFP for HCC are listed in Table 5. Cut off value of Talin-1 gene expression 9.5 (2-∆∆Ct) recorded an AUC of 85.7% with sensitivity of 93.3% and specificity of 80%. While cut off value of AFP 13.1 ng/mL showed an AUC of 65.4% with a sensitivity of 60% and specificity of 69.9%.
| Characteristics | HCC Group (N = 40) | Liver Cirrhosis Group (N = 30) | P Value |
| Age (years) | 60.92 ± 9.01 | 55.53 ± 9.64 | 0.02* |
| Gender Male Female | 31 (77.5%) 9 (22.5%) | 15 (50%) 15 (50%) | 0.02* |
| Residence Urban Rural | 25 (62.5%) 15 (37.5%) | 20 (66.67%) 10 (33.33%) | 0.72 |
| Occupation Farmer Non- farmer | 14 (35%) 26 (65%) | 4 (13.33%) 26 (86.67%) | 0.04* |
| Etiology Smoking Alcohol HCV HBV | 22 (55%) 1 (2.5%) 37 (92.5%) 2 (5%) | 11 (36.67%) 0 (0%) 22 (73.33%) 1 (3.33%) | 0.13 1 0.04* 1 |
| ALT (IU) | 34.97 (2.9-160) | 14.8 (19-87) | 0.000* |
| AST(IU) | 36.2 (28-137) | 12.7 (24-75) | 0.000* |
| ALP (IU) | 174.95 ± 121.36 | 167.17 ± 121.3 | 0.22 |
| Total Bilirubin mg/dl | 3.19 (0.6- 63) | 1.95 (0.5- 21) | 0.07 |
| Albumin g/dl | 2.62 ± 0.55 | 2.5 ± 0.77 | 0.45 |
| PT (seconds) | 17.29 ± 4.54 | 18.69 ± 10.9 | 0.56 |
| INR | 1.46 ± 0.31 | 1.6 ± 0.94 | 0.77 |
| Creatinine mg/dl | 2.12 ± 1.72 | 1.73 ± 1.3 | 0.33 |
Data are presented as mean ± standard deviation or median with minimum and maximum values, as appropriate. HCC: hepatocellular carcinoma; HCV: Hepatitis c virus; HBV: Hepatitis B virus; ALT:Alanine aminotransferase; AST:aspartate aminotransferase; ALP:alkaline phosphatase; PT:prothrombin time; INR:international normalized ratio *:Values statistically significant (at P< 0.05).
| Scoring System |
HCC Group (N= 40) |
Liver Cirrhosis Group (N = 30) |
P value | |||
| Child- Pugh score | - | - | - | |||
| Child A | 6 (15%) | 4 (13.33%) | 0.84 | |||
| Child B | 8 (20%) | 6 (20%) | 1 | |||
| Child C | 26 (65%) | 20 (66.67%) | 0.88 | |||
| MELD Score | - | - | - | |||
| Mean ± SD | 19.95 ± 9.29 | 19.1± 9.82 | 0.71 | |||
| Range | 6-44 | 7-45 | - | |||
| uMELD Score | - | - | - | |||
| Mean ± SD | 4.08 ± 1.1 | 4.03 ± 1.31 | 0.85 | |||
| Range | 2.6-7.1 | 2-6.6 | - | |||
| Characteristics | HCC Patients (N = 40) | Percentage (%) | |||
| Tumor size < 3 / 3-5 / > 5 cm | 9/ 15/ 16 | 22.5/ 37.5/ 40 | |||
| No. of nodules Single / 2 or more | 23 / 17 | 57.5 / 42.5 | |||
| Site of Tumor Right lobe/left lobe/both | 24/ 3 / 13 | 60/ 7.5/ 32.5 | |||
| ShapeRounded/oval /ill-defined | 23/12/5 | 57.5 / 30 / 12.5 | |||
| EchogenicityHyperechoic/hypoechoic/isoechoic | 24/13/3 | 60 / 32.5 / 7.5 | |||
| Halo sign | 38 | 95 | |||
| Metastasis | 1 | 2.5 | |||
| Portal Vein Invasion | 5 | 12.5 | |||
| Staging systems Okuda stage I / II /III CLIP stage I / II /III Tokyo stage Early (0- 4) Advanced (5 or more) VISUM stage I / II /III | 6 / 16 /18 4 /22/14 17 23 19 /13 /8 |
15/ 40 /45 10/ 55/ 35 42.5 57.5 47.5 / 32.5 /20 |
|||
| Parameters |
Group I HCC (N=40) |
Group II Cirrhosis (N=30) | Group III Control (N=30) | P value |
| Talin-1 Gene (2-∆∆CT) Mean± SD | 13.4 ±2.7 | 9.2 ±5.4 | 5 ±2.57 | P1=0.000* P2=0.000* P3=0.000* |
| AFP (ng/mL)Median (Range) | 37.47 (2.5-1000) | 10.55 (5.6 -51) | 5.69 (2.9-9) | P1=0.67 P2=0.004* P3=0.013* |
| Test | Cut-off value | Sensitivity% | Specificity% | PPV% | NPV% | AUC% | P-value |
| Talin-1 gene(2-∆∆CT) | 9.5 | 93.3 | 80 | 100 | 93.7 | 85.7 | 0.000* |
| AFP ng/ml | 13.1 | 60 | 69.9 | 100 | 71.4 | 65.4 | 0.018* |
4. DISCUSSION
Talin-1 can be considered as a protein involved in HCC progression. However, its explicit role is still unknown and its functional mechanism remains largely unclear [12, 34]. Assessment of gene expression in peripheral blood has the potential to inform on pathophysiological mechanisms and has emerged as a viable avenue for the identification of biomarkers [35, 36]. In our study, Talin-1 gene expression is significantly upregulated in the PBMCs of HCC patients in comparison to the cirrhotic and healthy subjects. In concordance with these results, Chen et al. [37] found that Talin-1 was highly expressed in HCC cells relative to non-cancer liver cells and had a role in tumor growth and metastasis. Similarly, Kanamori et al. (2011) [19] reported that Talin-1 expression levels in HCC nodules were significantly associated with their undifferentiation and a rapid recurrence after resection. Our results may adhere to the role of Talin-1 as oncogene-associated protein and along with the notion that its altered expression to be associated with human carcinogenesis. Talin-1 is a focal adhesion protein to mediate integrin interaction with extracellular matrix junctions, where it can bind to a variety of cell adhesion molecules and actin to induce cell cytoskeleton remodeling. Accordingly, an increase in Talin-1 expression means promotion of tumor cell adhesion and migration [38]. On the other hand, previous studies stated that Talin-1 is down-regulated in HCC liver tissues when compared with adjacent non-cancerous or control liver tissue [33]. Also, in contrast to ours, Chen et al. (2017) [12] observed that low Talin-1 expression was associated with HCC progression and poor prognosis. Moreover, Fang et al. (2014) [18] investigations showed that high levels of Talin-1 expression were correlated with low invasion and migration in human liver cancer cell lines; the suppression of Talin-1 promotes invasion and migration. In the same context, both Talin-1 and Talin-2 mRNA expression levels are related to tumorigenicity in human HCC. These molecules form important molecular targets for the diagnosis and/or treatment of HCC [39].
Clinical studies showed that the up-regulation of Talin-1 level occurred in tumors which were poorly differentiated tumors and angiogenesis or metastasis [21, 34]. However, among various populations, Talin-1 expression in HCC is still controversial. Khodeer et al. (2017) and Youns et al. (2013) [15, 40] found significantly higher levels of serum Talin-1 in HCC Egyptian patients in comparison to both cirrhotic and control groups. On the other hand, another study within the same population indicated significantly lower levels of serum Talin-1 in HCC patients than patients with cirrhosis and normal subjects [41]. Also, Talin-1 is upregulated in HCC in Japanese, down-regulated in Chinese. Significantly lower levels of Talin-1 protein and mRNA expression in HCC tissues than in the adjacent non-cancerous tissue were detected in Chinese people [18].
As regards the mechanisms by which Talin-1 promotes HCC progression and metastasis, recently, studies suggested that Talin-1 caused high expression of anti-apoptotic members of the B-cell lymphoma 2 (BCL2) family, and had a negative impact on the expression of apoptosis factors in the p53 network. Also, it was detected that Talin-1 promoted HCC metastasis by increasing Epithelial-Mesenchymal Transition (EMT) mesenchymal markers expression and inhibiting the expression of the epithelial molecules. In addition, Talin-1 contributed in the regulation of other biological behaviors related to HCC progression, including ion transport, membrane repolarization, cell growth, and adhesion [37]. Hu et al. (2017) [34] said that Talin1 could promote the proliferation of liver cancer cells and protect them from apoptosis by increasing the activity of the PI3K/Akt/NF-κB signaling pathway.
The pathogenesis of HCC is a multistep and multistage process involving environmental and genetic factors [ 42 ]. The molecular characterization of genetic alterations in the tumour cell population and their cellular composition, as well as the corresponding tumour microenvironment, can promote the development of predictive biomarkers for clinical practice [ 43 ]. Several studies suggest that Talin-1 can affect transcription via modulating interactions and signalling processes in focal adhesions at the cell membrane, which seem fundamentally involved in transcriptional regulation [ 44, 45].
During the last two decades, several studies have provided an increased understanding of the mechanism underlying HCC tumorigenesis, however, no method has been found to be suitable for the entire patient population due to the lack of specificity and sensitivity [ 46 ].
According to ROC analysis in this study, data revealed that Talin-1 gene expression could serve as a diagnostic marker for HCC. The cutoff value of 9.5 (2-∆∆Ct) exhibits sensitivity and specificity of 93.3% and 80% respectively. Additionally, we found that Talin-1 gene expression surpasses serum levels of AFP in the diagnosis of HCC. These results strongly support the previous Egyptian investigation on hepatocellular carcinoma [15, 40]. Also, we noted a statistically insignificant correlation between Talin-1 gene expression and Child score, MELD, uMELD, Tumor number, Tumor size, AFP, Okuda, CLIP, Tokyo score or VISUM staging systems; this was in agreement with Zhang et al. (2011) [32].
CONCLUSION
This study illustrated that Talin-1 gene expression is up-regulated in hepatocellular carcinoma patients. This expression has an evident diagnostic role with valuable sensitivity and specificity that transcend serum AFP for HCC detection.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The current study was approved by the Ethics and Research Committee of Benha faculty of Medicine, Benha University, Egypt.
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures were followed in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written informed consent was obtained from all the participants.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The author declares no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.


