All published articles of this journal are available on ScienceDirect.
The Utility of New Biomarker-based Predictive Model for Clinical Outcomes Among ST-elevation Myocardial Infarction Patients
Abstract
Aim:
To determine the discriminative potency of score to prognosticate poor clinical outcomes in ST-Segment Elevation Myocardial Infarction (STEMI) patients.
Methods:
From the entire population of STEMI (n=268), we enrolled 177 individuals with acute STEMI who underwent complete revascularization with primary Percutaneous Coronary Intervention (PCI). Clinical assessment, echocardiography, Doppler, and biomarkers’ measure were performed at baseline.
Results:
Combined endpoint (Major Cardiovascular Events - MACEs [composite of cardiovascular death, recurrent myocardial infarction, newly diagnosed Heart Failure] and hospitalization) was determined in 75 patients with acute STEMI population (40.6%). Newly onset heart failure (HF) was reported in 46 patients (26.0%), Cardiovascular (CV) death occurred in 12 patients (6.8%), MACEs were determined in 58 patients (32.8%), and recurrent hospitalization due to CV reasons was found in 17 (9.6%). The conventional risk predictive models were engineered by a combination of TIMI risk score +acute HF Killip class ≥ II + the levels of NT-pro brain natriuretic peptide > 300 pg / mL and troponin >0.05 ng/mL. We developed a new predictive model based on the presentation of T786С genotype of endothelial NO syntase gene (rs 2070744), А1166С in angiotensin-ІІ receptor-1 gene (rs5186) and serum levels of soluble suppressor tumorigenicity ≥35 pg/mL, vascular endothelial growth factor ≤172 pg/mL and macrophage inhibitory factor ≥2792.7 pg/mL. STEMI patients who had >5 score points demonstrated significantly worse prognosis than those who had ≤5 score points.
Conclusion:
Here we have reported that a new original predictive model is better than a conventional model in discriminative ability to predict combined clinical outcome in STEMI patients.
1. INTRODUCTION
During the last decade, the implementation of new technologies, such as early urgent Percutaneous Coronary Intervention (PCI), optical coherent tomography-guided PCI, early complete revascularization, implantation of new stents’ generation, and contemporary adjuvant therapy, has drawn a dramatic improvement in survival and decreased risk of Cardiovascular (CV) complications among patients with ST-Elevation Myocardial Infarction (STEMI) [1]. However, STEMI remains the main cause of premature CV death in the general population worldwide [2]. In-hospital mortality rate after successful complete revascularization (culprit artery and ischemic-related coronary arteries) in STEMI patients has dropped to 5-7% even in developed countries, and mortality rate within the first year after STEMI has been fluctuating around 14-17% [3]. A wide range of evidence supports that microvascular inflammation and small coronary artery occlusion after PCI are the most probable reasons for post-STEMI cardiac remodeling and one-year mortality [4-6]. Unfortunately, the clinical outcomes related have been poorly predicted by traditional risk scores including TIMI and GRACE that shape sustainable scientific interest in novel personified predictive models [7].
Currently there are numerous risk predictive tools based on measurement of circulating levels of the biomarkers, which represent different pathobiological axes of STEMI natural evolution and have been developed to stratify STEMI patients at high CV risk [8-10]. Several studies have assessed the combination of circulating biological markers (i.e., troponin T and -I, soluble Suppressor Tumorigenicity-2 (sST2), natriuretic peptides, galectin-3, metalloproteinases, growth / differential factor-15, proadrenomedullin, Macrophage Inhibitory Factor-1 (MIF), C-reactive protein, creatinine, lipid profile) with multiple markers risk score for STEMI prognostication [11-13]. Therefore, previous studies have unleashed that numerous genetic biomarkers (Single Nucleotide Polymorphism (SNP) in the promoter region of endothelial NO synthase, aldosterone synthase (CYP11B2), and angiotensin-II receptor-1 genes, SNP Val66Met in Brain-Derived Neurotrophic Factor BDNF gene, SNP Lys198Asn in endothelin [EDN]-1 gene, Vascular Endothelial Growth Factor (VEGF) gene) well corresponded to post-STEMI adverse cardiac remodeling, early stent thrombosis, restenosis, microvascular obstruction and no-reflow phenomenon after PCI [14-18].
However, the discriminative potencies of both circulating biomarkers and genetic polymorphisms to identify patients at increased risk of CV death or Heart Failure (HF) were not deeply investigated among STEMI patients who underwent complete early PCI in a contest of getting additive value beyond established clinical predictors and / or contemporary risk scores (the TIMI and GRACE Risk Score for STEMI) [19]. The aim of the study was to determine the discriminative potency of new original predictive scores to prognosticate poor clinical outcomes in STEMI patients treated with complete revascularization.
2. METHODS
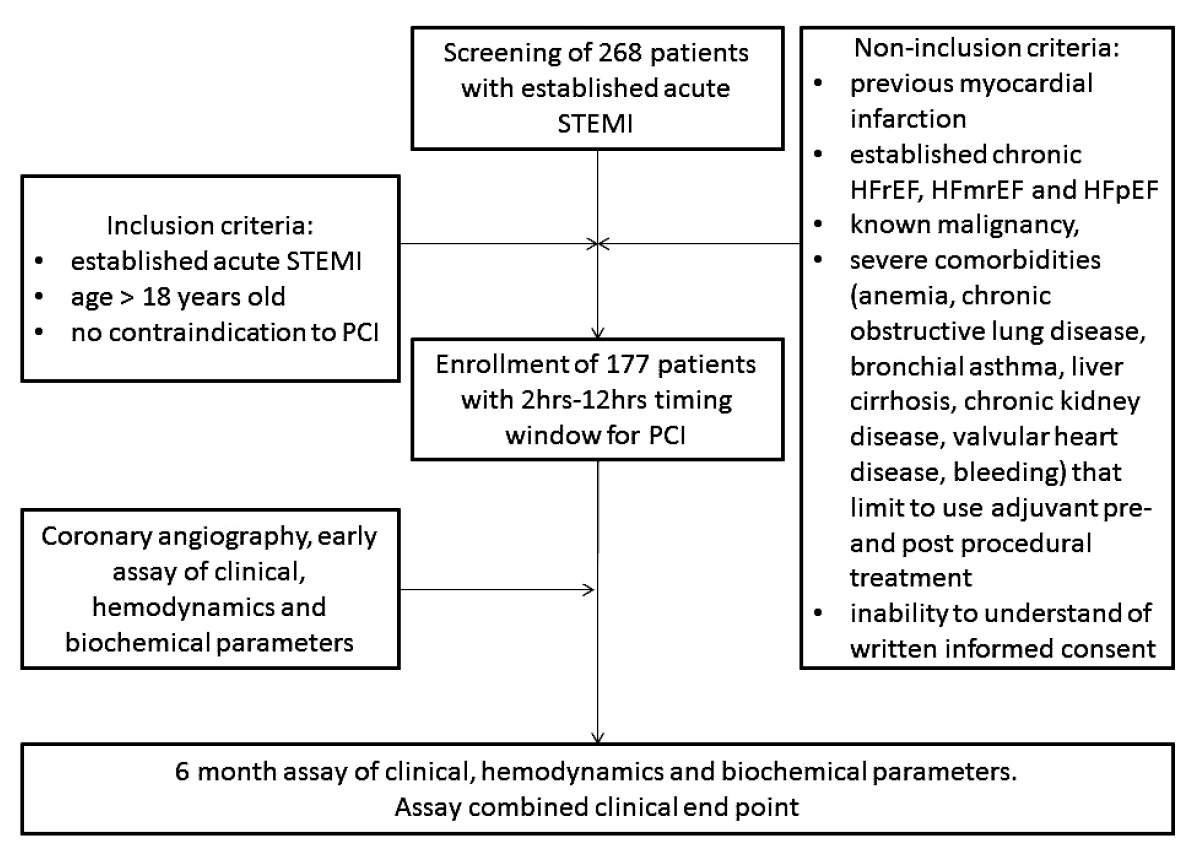
A total of 268 patients with confirmed acute STEMI were analyzed for participation in the study (Fig. 1). From the entire population of STEMI (n=268), according to inclusion and non-inclusion criteria, we enrolled 177 individuals with acute STEMI who were admitted to the intensive care unit of “L.T.Malaya TNI NAMSU” (Kharkiv, Ukraine) within a given period from 2016 January to 2019 June. Acute STEMI was diagnosed according to ECS Guidelines (2017) [20].
The inclusion criteria involved acute STEMI, age > 18 years old, a lack of contraindications to PCI, and a written informed consent to participate in the study. Exclusion criteria comprised of previous myocardial infarction, established chronic heart failure, severe comorbidities (anemia, chronic obstructive lung disease, bronchial asthma, liver cirrhosis, chronic kidney disease with declined glomerular filtration rate < 35 mL/min×1.73m2, valvular heart disease, bleeding), known malignancy and pregnancy, as well as the inability to understand the informed consent.
Primary PCI with bare-metal stent (COMMANDER, “Alvimedica”, Turkey) implantation was performed in 104 patients within 6-12 hours after initial acute STEMI confirmation in the V.T. Zaytsev Institute of General and Emergency Surgery NAMSU (Kharkiv, Ukraine). Systemic thrombolysis (tPA tenecteplase i.v. bolus per conventional protocol) was carried out in 31 STEMI patients prior to PCI. All acute STEMI patients received adjuvant treatment due to current ESC recommendations [20]. TIMI III blood flow through the culprit artery was determined for every re-perfused patient with acute STEMI (Fig. 2).
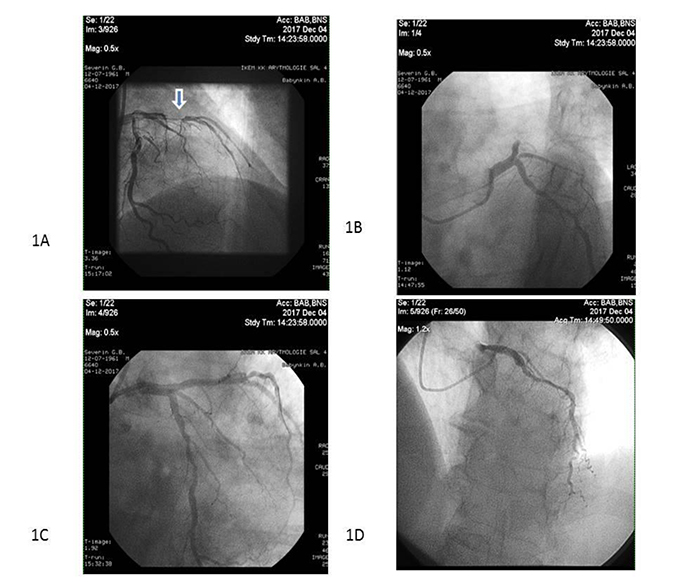
Note: Arrow indicates the culprit artery.
2.1. Ethical Declaration
All procedures performed in the study involving human participants were in accordance with the ethical standards and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards and approved by the local ethics committee (Protocol Nº8, August 29th 2016). Written voluntary informed consent was obtained from each patient before entering the study.
2.2. Sample Size Calculation
The sample size was calculated through the effect size estimation (0.99), the type of the present study, providing study power of 80% and type I error 5%, STEMI in-hospital mortality of 7.5% and one-year mortality of 14% [21]. The sample size was supposed to be comprised of at least 170 individuals.
2.2.1. Coronary Angiography
Conventional coronary angiography was performed immediately after admission of the patients to the hospital using Digital X-Ray system “Integris Allura” (Philips Healthcare, Best, The Netherlands) and managed by radial or femoral vascular access. Coronary arteries were visualized with two-to-three orthogonal projections per conventional protocol. The number of views obtained was decided by the operator depending on coronary anatomy. The main coronary arteries were left main coronary artery, left anterior descending branch, left circumflex branch, right main coronary artery and right coronary descending branch. In this study, the contrast “Ultravist-370” (Baier Pharma GmbH, Germany) and automatic contrast injector were used. The contrast amount used in coronary angiography in each injection was 8 - 10 mL at 4 mL/s for the left coronary artery and 6 mL at 3 mL/s for the right coronary artery (radiation exposure 20 to 35 mGycm). After coronary angiography, two experienced interventional cardiologists discussed the captures and filled in the final report of the results of the procedure after reaching consensus.
2.3. Determination of Risk Factors and Comorbidities
Hypercholesterolemia (HCE) was diagnosed if Total Cholesterol (TC) level was above 5.2 mmol/L, and/or Low-Density Lipoprotein cholesterol (LDL) level was above 3.0 mmol/L, and/or level of triglycerides (ТG) was above 1.7 mmol/L according to the European Cardiology Society dyslipidaemia guideline (2016) [22]. Hypertension was diagnosed if Systolic Blood Pressure (SBP) was >140 mm Hg, and/or Diastolic Blood Pressure (DBP) >90 mm Hg according to the European guideline on diagnostics and treatment of arterial hypertension (2018) [23]. Heart failure was diagnosed according to ESC guidelines for the diagnosis and treatment of acute and chronic heart failure (2016) [24]. Positive smoking history was defined as having smoked daily or occasionally in the past.
2.4. Transthoracic Echocardiography and Doppler
Transthoracic Echocardiography was performed on “Aplio 500” (TUS-A500) TOSHIBA MEDICAL SYSTEMS CORPORATION with usage of 3.5 MHz phase probe at discharge and at 6-month observation period. Left Ventricular (LV) End Diastolic Volume (LVEDV), LV end systolic volume (LV ESV), LV Ejection Fraction (LVEF) measuring were done according to Simpson's method per contemporary recommendation. Left Atrial Diameter (LAD) and Left Atrial Volume (LAV) were determined according to the contemporary protocol [25]. LV global longitudinal strain (e`) and early transmitral velocity (E) were measured by tissue Doppler imaging technique and impulse transmitral Doppler regime at baseline per protocol.
2.5. Determination of STEMI Prognosis
We used the TIMI score and the GRACE score to validate prognostic capacity after STEMI [26, 27]
2.5.1. SYNTAX Score Determination
SYNTAX score (SS) was used to assess the severity of coronary atherosclerotic lesions and it was calculated by experienced interventional cardiologist accordingly [28].
2.6. Determination of Endpoint
The endpoint was determined as a combined event including CV death, recurrent angina / myocardial infarction, newly diagnosed heart failure and hospitalization for 6 months after PCI. CV death was ascertained by personal or phone contact with the family doctor or the hospital where the patient died. The diagnosis of recurrent angina required the presence of clinical signs/symptoms or electrocardiographic changes. Hospitalization was ascertained by direct contact or phone call to the hospital reception where the patient was admitted. Discharge report or autopsy report was obligatorily reviewed before endpoint determination.
2.7. Calculation of Glomerular Filtration Rate
Glomerular Filtration Rate (GFR) was calculated by the CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration) equation [29].
2.8. Blood Samples
Blood samples were drawn immediately before PCI at baseline and at six months of the post-PCI period. Blood samples were centrifuged, serum was isolated within 30 minutes of sample acquisition, and then they were stored in plastic tubes and frozen at -70 C until being shipped to the laboratory of immunochemical and molecular-genetic researches of GI “L.T.Malaya TNI NAMSU”.
2.9. Circulating Biomarkers Measurements
Troponin I (Tn I) level was measured by chemoluminescent immunoassay (Humalyzer 2000, HUMAN GmbH, Germany) according to the manufacturers’ recommendations. The average of Tn I level was 0.5-50 ng/mL.
Total Creatine Kinase (CK) and CK MB-fraction (CK-MB) were analysed using immunoinhibition method on quantitative immunoassay analyser Humalyser 2000 (HUMAN GmbH, Germany) according to the manufacturers’ recommendations.
Total Cholesterol (TC), Low-Density Lipoprotein (LDL) cholesterol, high-density lipoprotein (HDL) cholesterol and Triglycerides (TG) were measured by direct enzymatic method (Roche P800 analyser, Basel, Switzerland).
Fasting glucose level was measured by double-antibody sandwich immunoassay (Elecsys 1010 analyser, F. Hoffmann-La Roche Diagnostics, Mannheim, Germany).
N-terminal fragment of brain natriuretic peptide (NT-proBNP) was measured by a commercially available standard kit (R&D Systems GmbH, Wiesbaden-Nordenstadt, Germany). The NT-proBNP level average was 10-12000 pg/mL.
Soluble suppressor tumorigenicity-2 (sST2) levels were measured by commercially available standard kit Presage ST2 Assay (Critical Diagnostics, San Diego, Ca, USA). The sST2 level average was 0-200 ng/mL.
Macrophage Inhibitory Factor-1 (MIF) levels were measured using Humalyzer 2000 (HUMAN GmbH, Germany) by the enzyme-linked immunoassay method (RayBio® Human MIF ELISA KIT, USA). The MIF level average was from 0 to 6000 pg/mL.
The levels of VEGF-A were measured with a commercial kit for ELISA (IBL International GMBH, Germany). The VEGF-A level average was from 0 to 1000 pg/mL.
The intra-assay and inter-assay coefficients of variation for all biomarkers were <5%.
2.10. Genetic Biomarkers Determination
The DNA extraction was performed according to the protocol for a commercial set «TacMan TMSNP Genotyping Assays» (Thermo Fisher Scientific Assay IDC_11592758_1). The assessment of allelic states of SNP studied was performed using Real-Time (RT) Polymerase Chain Reaction (PCR). RT PCR was performed on CFX96 thermocycler (BioRad, USA) using an allelic discrimination test.
SNP T786C (rs2070744) in eNOS gene determination
The primers’ sequences were 5’-ACCAGGGCATCA AGCTCTTC-3’, 5’-GCAGGTCAGCA GAGAGACTAG-3’, rs2070744-C: 5’-VIC-AGGGTCAGCC GGCCAG-BHQ1-3’, rs2070744-T: 5’-FAM-AGGGTCAGCCAGCCAGBHQ1-3’.
SNP Val66Met (rs6265) in BDNF gene determination
Primers that we used in BDNF val66met (rs6265) polymorphism assay were as follows: CCTACAGTTCCACC AGGTGAGAAGAGTG (forward), TCATGGACATGTTTG CAGCATCTAGGTA (reverse).
SNP Т344С (rs1799998) in CYP11B2 gene determination
CYP11B2 primer sequences were TTTATCTTATCGT GAGATGAGAGGG (forward), GCCTTGGATTCTTTTAA TAGACTTT (reverse).
SNP А1166С (rs5186) in AGT2R1 gene determination
AGT2R1 primer sequences were TGCAGCACTTCAC TACCAAATGAGC (forward), TTAGCTACTTTTCAGAAT TGAAGGA (reverse).
SNP Lys198Asn (rs5370) in EDN-1 gene determination
Lys198Asn primer sequences were TTCATGATCCCA AGCTGAAAGGCAA (forward) and CCCTCCAGAGAGCG TTATGTGACCC (reverse).
2.11. Statistics
Statistical analyses were performed using SPSS for Windows v. 23 (IBM, Chicago, USA). Continuous variables are presented as mean (M) ± Standard Deviation (SD) and mean and 95% Confidence Interval (CI) when they were normally distributed, or in median and interquartile range if otherwise. Categorical variables are presented as frequencies and percentages. Mann-Whitney and Wald-Wolfowitz’s criteria were used for intergroup differences and quantitative values. The qualitative variables are expressed as percentages, and were analysed by the χ2 test and exact Fisher test. Allele frequencies were estimated, and all polymorphisms were tested for Hardy–Weinberg Equilibrium. Correlations between single nucleotide gene polymorphisms, angiographic characteristics, hemodynamic performances, and biomarkers were received using rang correlation r Spearmen test. We performed univariate and multiple variate log-regression analysis to determine variables that predict the combined clinical endpoint. Beta coefficient, Standard Errors (SE), Odds Ratio (OR), 95% Confidence Interval (CI) for each factor were estimated. The Receiver Operating Characteristic (ROC) curve was performed for the detection of a well-balanced cut-off of biomarkers’ concentrations, TIMI score index and SYNTAX score index. Area Under Curve (AUC), Sensitivity (Se), Specificity (Sp), Positive (PPV) and Negative (NPV) Predictive Values, positive (PLR) and negative (NLR) Likelihood Ratios were calculated for each model. Predictive models were compared with C-statistic. The Integrated Discrimination Indices (IDI) and Net-Reclassification Improvement (NRI) were utilized for prediction performance analyses. Prognostic Coefficient (PC) was calculated by the equation: PC=100log (frequency of combined endpoint / frequency of free combined endpoint). Survival analysis for clinical outcomes was performed using Kaplan–Meyer curves and the log-rank test. All differences were considered statistically significant with 2-tailed P<0.05.
3. RESULTS
Combined endpoint (MACEs [composite of CV death, recurrent MI, newly diagnosed HF] and hospitalization) was determined in 75 patients with acute STEMI population (40.6%). Newly onset heart failure was reported in 46 patients (26.0%), CV death occurred in 12 patients (6.8%), MACEs were determined in 58 patients (32.8%), and recurrent hospitalization due to CV reasons was found in 17 (9.6%).
The basic characteristics of the entire patient population are reported in Table 1. The STEMI population was constructed from predominantly male (78.5%), aged 46 years to 74 years (average age was 61.73 years), having severe CV risk factors, such as hypertension (82.5%), diabetes mellitus (24.9%), smoking (47.5%), hypercholesterolemia (59.3%), abdominal obesity (39.0%). Stable and unstable angina pectoris prior to STEMI were found in 60.5% and 37.9% patients, respectively. Hemodynamic characteristics showed that the average of LVEF and E/e` ratios were 51.82% and 11.6 units, respectively. Patients who met combined clinical outcomes did not differ from those who had no clinical outcomes in average age, gender, CV risk factors, and hemodynamics except left atrial volume (P=0.021), left atrial dimension (P=0.045), and E/e`ratio (P=0.042). Additionally, there were no significant differences in concomitant medications between both patients’ cohorts.
| Variables | Entire STEMI Population (n=177) | STEMI Patients | P value | |
|---|---|---|---|---|
| Who Met Combined Clinical Outcomes (n=75) | Who were free of Combined Clinical Outcomes (n=102) | |||
| Demographic, comorbidities and CV risk factors | ||||
| Age, years (SD) | 61.73±9.44 | 59.8±8.52 | 58.27±6,75 | 0.185 |
| Male, n (%) | 139 (78.5) | 56 (74.7) | 84 (82.4) | 0.214 |
| Female, n (%) | 38 (21.5) | 19 (25.3) | 18 (17.6) | |
| Hypertension, n (%) | 146 (82.5) | 63 (84.0) | 83(81.4) | 0.653 |
| T2DM, n (%) | 44 (24.9) | 23(30.7) | 21(20.6) | 0.126 |
| Smoking, n (%) | 84 (47.5) | 39(52.0) | 45(44.1) | 0.300 |
| HCE, n (%) | 105 (59.3) | 46(61.9) | 59(57.8) | 0.584 |
| Obesity (BMI>30 кг/м2), n (%) | 69 (39.0) | 34(45.3) | 35(34.3) | 0.139 |
| Stable CAD prior to STEMI, n (%) | 107 (60.5) | 49(65.3) | 58(56.9) | 0.260 |
| Unstable angina prior to STEMI, n (%) | 67 (37.9) | 33 (44.0) | 34 (33.3) | 0.149 |
| Concomitant medications | ||||
| Beta-blockers, n (%) | 177 (100) | 75 (100) | 102 (100) | 0.99 |
| ACEI / ARBs, n (%) | 165 (93.2) | 72 (96.0) | 93 (91.2) | 0.46 |
| Clopidogrel /Ticagrelor, n (%) | 172 (97.2) | 73 (97.3) | 99 (97.1) | 0.94 |
| Statins, n (%) | 177 (100) | 75 (100) | 102 (100) | 0.99 |
| MCRAs, n (%) | 128 (72.3) | 53 (70.7) | 75 (73.5) | 0.89 |
| Hemodynamic | ||||
| HR, per min | 76.89±15.52 | 77.72±14.73 | 74.45±17.52 | 0.192 |
| SBP, mmHg | 134.87±25.83 | 136.76±28.27 | 130.94±25.83 | 0.157 |
| DBP, mmHg | 80.62±12.53 | 81.36±12.56 | 78.43±12.28 | 0.122 |
| LV EDV, ml | 136.71±37.67 | 136.06±38.90 | 138.38±33.63 | 0.672 |
| LV ESV, ml | 64.76±28.32 | 65.29±29.27 | 63.04±25.20 | 0.430 |
| LA, cm | 4.10±0.51 | 4.22±0.49 | 4.06±0.56 | 0.045 |
| LAV, ml | 56.2±7.91 | 57.8±10.7 | 54.2±9.8 | 0.021 |
| LV EF, % | 51.82±10.53 | 51.23±10.62 | 53.55±10.13 | 0.142 |
| E/e`ratio, unit | 11.6±4.28 | 12.89±5.34 | 11.31±4.86 | 0.042 |
The baseline characteristics of biomarkers in STEMI patients are given in Table 2. The STEMI patients who met combined clinical outcomes had significantly higher levels of biomarkers of necrosis (peak troponin I, peak CK-MB), biomechanical stress (NT-proBNP), inflammation and fibrosis (MIF, sST2), angiogenesis and endothelial function (VEGF-A), than STEMI individuals without poor outcomes. Therefore, the serum levels of low-density lipoprotein cholesterol were sufficiently lowered in the patients who did not meet combined clinical outcomes than in patients who did (P=0.031). There were no significant differences between both the cohorts in serum levels of total cholesterol, triglycerides, high-density lipoprotein cholesterol, as well as estimated GFR values were similar in both cohorts.
| Variables | Entire STEMI Population (n=177) | STEMI Patients | P value | |
| Who met Combined Clinical Outcomes (n=75) | Who were free of Combined Clinical Outcomes (n=102) | |||
| Circulating Biomarkers | ||||
| PeakTnI, ng/mL | 17.72 [6.34-77.23] |
18.46 [8.55-99.45] |
13.18 [5.97-68.5] |
0.038 |
| Peak CK-MB,U/L | 103.3 [44.9-28.95] |
156.9 [123.3–359.0] |
81.8 [34,0–153,9] |
0.004 |
| NT-proBNP, pg/mL | 246.81 [26.78 – 610.97] |
415.12 [74.45-1305,42] |
202.43 [54.48-802.60] |
0.001 |
| sST2, ng/mL | 45.81 [32.23-102.47] |
63.72 [35.99-134.53] |
44.74 [28.25-77.32] |
0.018 |
| VEGF-A, pg/mL | 160.33 [83.82–299.62] |
229.62 [108.86–379.00] |
103.79 [69.80–157.60] |
0.028 |
| MIF, pg/mL | 2582.80; [1308.40 - 4122.20] | 3954.00 [3076.30-4964.30] |
1277.85 [556.70-1931.80] |
0.001 |
| Total cholesterol, mmol / L | 4.82 [3.95-5.63] | 4.93 [3.98-5.74] | 5.08 [4.10-5.79] | 0.602 |
| TG, mmol/L | 1.53 [1.17-2.02] | 1.63 [1.19-2.06] | 1.45 [1.13-1.91] | 0.184 |
| HDL-cholesterol, mmol / L | 1.12 [0.92-1.28] | 1.11 [0.90-1.31] | 1.01[0.90-1.20] | 0.359 |
| LDL cholesterol, mmol / L | 3.00 [2.03-3.63] | 3.24 [2.11-3.67] | 2.91 [2.07-3.99] | 0.031 |
| GFR, ml/min | 104.67±27.56 | 103.68±27.77 | 107.50±26.96 | 0.389 |
| Genetic biomarkers (SNP) | ||||
| T786C haplotypes in eNOS gene (rs2070744), n (%) | ТТ: 73 (41.2%) ТС: 64 (36.1%) СС: 40 (22.6%) |
ТТ: 21 (28.0%) ТС: 20 (26.7%) СС: 34 (45.3%) |
ТТ: 22 (21.6%) ТС: 58 (56.9%) СС: 22 (21.6%) |
0.0001 |
| Lys198Asn haplotypes in EDN-1 gene (rs5370), n (%) |
LysLys: 74 (41.8%)
LysAsn+AsnAs: 103 (58.2%)* |
LysLys: 30 (40%)
LysAsn+AsnAs: 45 (60%)* |
LysLys: 44 (43.1%)
LysAsn+AsnAs: 58 (56.9%)* |
χ 2 =0.17, р=0.676 |
| Т344С haplotypes in CYP11B2 gene (rs1799998), n (%) |
ТТ: 58 (32.8%)
ТС: 88 (49.7%) СС: 31 (17.5%) |
ТТ: 27 (36%) ТС: 40 (53.3%) СС: 8 (10.7%) |
ТТ: 31 (30.4%) ТС: 48 (47.1%) СС: 23 (22.5%) |
0.326 |
| Val66Met haplotypes in BDNF gene (rs6265), n (%) |
ValVal: 45 (25.4%)
ValMet+MetMet: 132 (74.6%)* |
ValVal: 13 (17.4%) ValMet+MetMet: 62 (82.6%)* |
ValVal: 32 (31.4%) ValMet+MetMet: 70 (68.6%)* |
χ2=4.49, 0.034 |
| А1166С haplotypes in ATІІR1 gene (rs5186), n (%) |
АА: 118 (66.7%),
АС+СС: 59 (33.3%) |
АА: 41 (54.7%), АС+СС: 34 (58.7%) |
АА: 77 (75,5%), АС+СС: 25 (24.5%) |
χ2=8.43, 0.0037 |
There were significant differences between both cohorts of STEMI patients in the frequencies of SNPs of T786C in eNOS gene (rs2070744), Val66Met in BDNF gene (rs6265), and А1166С in ATІІR1 gene (rs5186), whereas the frequencies of Lys198Asn in EDN-1 gene (rs5370) and Т344С in CYP11B2 gene (rs1799998) were similar in both patients’ cohorts.
STEMI patients who met combined clinical outcomes demonstrated a higher TIMI risk score index that those who did not (Table 3). Although the total SYNTAX score index was similar in both patients’ cohorts, high severity of coronary atherosclerotic lesions (>32 points) was rather found frequently in STEMI patients with combined clinical outcomes than patients who did not meet these outcomes (P=0.011). The values of GRACE score index did not differ in both patients’ cohorts. Overall, MI due to LAD lesions was found frequently in patients with poor clinical outcomes than in patients without combined endpoint, whereas proportions of the patients with RCA type lesions and LCX lesions in both cohorts were similar (P=0.181 and P=0.808, respectively). Additionally, significant differences were not found between patients’ cohorts in a number of injured coronary arteries, frequencies of injury regarding LAD, RAD, and LCX except LCA.
Receive operation curve analysis for well-balanced levels of circulating biomarkers and risk scores
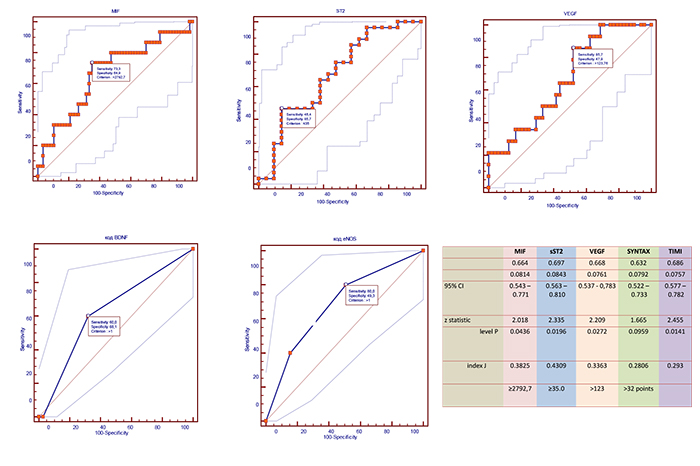
Using ROC analysis, we found that circulating levels of MIF≥2792.7 pg/mL, VEGF >123 pg/mL, sST2 ≥35 pg/mL, as well as SYNTAX score > 32 points, and TIMI score > 6 points were optimal having balanced sensitivity and specificity to predict combined clinical endpoint (Fig. 3).
| Variables | Entire STEMI Population (n=177) | STEMI Patients | P value | |
|---|---|---|---|---|
| Who Met Combined Clinical Outcomes (n=75) | Who were free of Combined Clinical Outcomes (n=102) | |||
| STEMI Risk Scoring | ||||
| TIMI risk score, point | 6 [4-7] | 8 [5-9] | 6 [4-8] | 0.046 |
| Total SYNTAX score, point | 28.7±6.15 | 27.54±6.41 | 25.65±8.82 | 0.134 |
| >32 points, n (%) | 76 (42.9) | 41 (54.6) | 35 (34.3) | 0.011 |
| 22 - 32 points, n (%) | 79 (44.6) | 38 (50.7) | 41 (40.2) | 0.167 |
| ≤22 points, n (%) | 22 (12.4) | 10 (13.3) | 12 (11.8) | 0.765 |
| Total GRACE Score, points | 150 (120-172) | 143 (117-170) | 152 (119-176) | 0.294 |
| STEMI Localization | ||||
| LAD lesions, n (%) | 77 (43.5%) | 52 (69.3) | 25 (24.5) | 0.001 |
| RCA type lesions, n (%) | 70 (39.5%) | 34 (45.3) | 36 (35.3) | 0.181 |
| LCX lesions, n (%) | 20 (11.3%) | 9 (12.0) | 11 (10.8) | 0.804 |
| Number of Coronary Arteries Injured | ||||
| One artery, n (%) | 53 (29.9%) | 27 (36.0) | 26 (25.5) | 0.134 |
| Two and more arteries, n (%) | 103 (58.2%) | 49 (65.3) | 54 (52.9) | 0.100 |
| LAD, n (%) | 47 (26.6%) | 21 (28.0) | 26 (25.5) | 0.710 |
| RCA, n (%) | 41 (23.2%) | 19 (25.3) | 22 (21.5) | 0.554 |
| LCX, n (%) | 20 (11.3%) | 9 (12.0) | 11 (10.8) | 0.804 |
| LCA, n (%) | 12 (6.8%) | 9 (12.0) | 3 (2.9) | 0.018 |
Univariate and multivariate linear regressions for 6-month combined endpoint after STEMI
The univariate log regression (stepwise) analysis has shown that С786С genotype of eNOS gene (rs2070744), Val66Met in BDNF gene (rs6265), А1166С in ATІІR1 gene (rs5186), MIF≥2792.7 pg/mL, VEGF ≤172 pg/mL, sST2 ≥35 pg/mL, SYNTAX score >32 points, TIMI score >6 points, abdominal obesity, unstable angina prior to acute STEMI, NT-proBNP > 300 pg/mL were found as significant predictors for combined clinical endpoint (Table 4). Other variables did not embed into multivariate log regressive analysis due to P>0.01. Further multivariate log regressive analysis has revealed that С786С genotype of eNOS gene (rs2070744), Val66Met in BDNF gene (rs6265), А1166С in ATІІR1 gene (rs5186), MIF≥2792.7 pg/mL, VEGF ≤172 pg/mL, sST2 ≥35 pg/mL, SYNTAX score >32 points, and TIMI score >6 points remained independent significant predictors for combined endpoint. After adjustment for severity of coronary atherosclerosis, С786С genotype of eNOS gene (rs2070744), Val66Met in BDNF gene (rs6265), А1166С in ATІІR1 gene (rs5186), MIF≥2792.7 pg/mL, VEGF ≤172 pg/mL, sST2 ≥35 pg/mL unleashed their predictive potency compared with the standard model and each other.
| Data | Depending Variable: Combined end Point | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Linear Regressive Analysis | Multivariate Linear Regressive Analysis | |||||||
| β-coefficient | OR | 95% CІ | Р | β-coefficient | OR | 95% CІ | Р | |
| Standard (reference) model | 1.44380 | 1.9620 | 1.1520 – 8.3644 | 0.0230 | 1.27480 | 1.6740 | 1.12710 – 6.9520 | 0.0460 |
| С786С genotype of eNOS gene (rs2070744) (present vs. absent) | 1.58366 | 4.8728 | 1.4093 – 16.8481 | 0.0123 | 1.57342 | 4.8231 | 1.5349 – 15.1552 | 0.0071 |
| Val66Met in BDNF gene (rs6265) (present vs. absent) | 0.74033 | 2.0966 | 1.0945 – 4.4990 | 0.0470 | 0.68301 | 1.9798 | 1.1545 – 4.1065 | 0.0395 |
| MIF≥2792.7 pg/mL | 1.2944 | 1.1253 | 1.1137 - 1.2722 | 0.0489 | 1.2527 | 1.1162 | 1.0965 - 1.2144 | 0.0488 |
| А1166С in ATІІR1 gene (rs5186) (present vs. absent) | 1.1814 | 1.1433 | 1.0850 – 2.2100 | 0.0414 | 1.1522 | 1.1243 | 1.0663 – 1.9811 | 0.0466 |
| VEGF ≤123 pg/mL vs. >123 pg/mL | 1.1544 | 1.1537 | 1.0766 – 2.0132 | 0.0301 | 1.1544 | 1.1244 | 1.0531 –1.8832 | 0.0426 |
| sST2 ≥35 pg/mL vs. < 35 pg/mL | 1.49352 | 1.1625 | 1.0944 - 1.2617 | 0.001 | 1.2832 | 1.1224 | 1.1164-1.1626 | 0.002 |
| SYNTAX score >32 points vs. ≤ 32 points | 1.17560 | 1.9428 | 1.2493 – 3.5422 | 0.0244 | 1.41380 | 1.6844 | 1.1830 – 2.3655 | 0.0234 |
| TIMI score >6 points vs. ≤ 6 points | 1.37250 | 1.8970 | 0.9720 – 2.880 | 0.0410 | 1.17280 | 1.0940 | 1.010 – 1.3240 | 0.0420 |
| Smoking (present vs. absent) | 0.51264 | 1.6697 | 0.3756 – 7.4222 | 0.5006 | - | - | - | - |
| T2DM (present vs. absent) | 0.35065 | 0.7042 | 0.2961 – 1.6748 | 0.4276 | - | - | - | - |
| Abdominal obesity (present vs. absent) | 1.12320 | 2.1448 | 0.4607 – 3.8995 | 0.0383 | 1.02 | 1.9560 | 0.0774 – 3.4539 | 0.0526 |
| Killip class of acute HF before PCI (II-III class vs I class) | 0.14908 | 0.8615 | 0.0713 – 4.3338 | 0.8565 | - | - | - | - |
| Stable CAD prior to STEMI (present vs. absent) | 0.43968 | 1.5522 | 0.3988 – 6.0419 | 0.5260 | - | - | - | - |
| Unstable angina prior to acute STEMI (present vs. absent) | 0.78264 | 2.3177 | 1.0611 – 4.1522 | 0.0462 | 0.71551 | 1.2317 | 0.9815 – 4.1772 | 0.1622 |
| Multiple coronary vessel injury vs. single coronary vessel injury | 0.22359 | 0.7996 | 0.1766 – 1.2622 | 0.3370 | - | - | - | - |
| E/e` ratio, > 15 units vs. ≤15 units | 0.35360 | 0.9160 | 1.0136 – 1.1630 | 0.0870 | - | - | - | - |
| LDL cholesterol, per 0.5 mmol/L | 0.72550 | 1.4271 | 0.9388 – 3.229 | 0.6630 | - | - | - | - |
| NT-proBNP > 300 pg/mL vs. ≤300 pg/mL | 1.18440 | 1.7044 | 1.0633 – 2.954 | 0.03420 | 1.17230 | 1.0144 | 1.0330 – 1.1422 | 0.0620 |
| Peak TnI, >0.05 ng/mL vs. ≤ 0.05 ng/mL | 0.97510 | 1.1774 | 1.0814 – 1.302 | 0.0467 | 0.9880 | 1.1034 | 1.0024 – 1.1852 | 0.0710 |
| Peak CK-MB, per 50 U/L | 0.47640 | 1.0254 | 1.0180 – 1.104 | 0.4820 | - | - | - | - |
| Hypercholesterolemia (present vs. absent) | 0.4582 | 0.8848 | 0.6638 – 1.1255 | 0.6388 | - | - | - | - |
3.1. Comparisons of Predictive Values for Different Models
We performed the face-to-face comparisons of different predictive models including standard model (combination of acute HF Killip class ≥ II + NT-proBNP > 300 pg / mL + troponin >0.05 ng/mL) in terms of sensitivities, specificities, positive and negative predictive values and likelihood ratios (Table 5). We found that two models based on four biomarkers (Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL; Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + sST2 ≥35 pg/mL), two models based on five biomarkers (Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + С786С genotype of eNOS gene; Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + sST2 ≥35 pg/mL), and one model based on six biomarkers (Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + С786С genotype of eNOS gene + sST2 ≥35 pg/mL) have demonstrated optimal balance between sensitivity (80% and more for all cases) and specificity (66% and more for all cases) with notable levels of positive (90% and more for all cases) and negative (40% and more for all cases) predictive values.
| Predictive Models | AUC | CI | P | Se, % | Sp, % | PPV, % | NPV, % | PLR, % | NLR, % |
|---|---|---|---|---|---|---|---|---|---|
| Standard model | 0.547 | 0.450 - 0.620 | 0,4862 | 75.0 | 50.0 | 100 | 20 | 1.5 | 0.5 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL | 0.702 | 0.581 – 0.805 | 0,0243 | 71.6 | 33.3 | 86 | 20 | 1.07 | 0.86 |
| Val66Met in BDNF gene + А1166С in ATІІR1 gene | 0.711 | 0.602 – 0.804 | 0.0250 | 61.6 | 50.0 | 87 | 30 | 1.22 | 0.78 |
| Val66Met in BDNF gene + VEGF ≤123 pg/mL | 0.694 | 0.563 – 0.806 | 0.0284 | 82.8 | 75.0 | 100 | 23 | 3.23 | 0.73 |
| Val66Met in BDNF gene + С786С genotype of eNOS gene | 0.712 | 0.605 – 0.805 | 0.0208 | 81.6 | 33.3 | 100 | 20 | 1.5 | 0.5 |
| Val66Met in BDNF gene + sST2≥35 pg/mL | 0.782 | 0.656 – 0.879 | 0.0073 | 82.1 | 33.3 | 96 | 39 | 1.22 | 0.55 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene | 0.776 | 0.661 – 0.866 | 0.0090 | 86.2 | 83.3 | 98 | 36 | 5.06 | 0.17 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + VEGF≤172 pg/mL | 0.768 | 0.625 – 0.877 | 0.0249 | 79.1 | 50.0 | 92 | 25 | 1.58 | 0.42 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL ≥2792.7 pg/mL + С786С genotype of eNOS gene | 0.781 | 0.667 – 0.870 | 0,0144 | 80.6 | 45.0 | 95 | 7 | 1.45 | 0.44 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + sST2≥35 pg/mL | 0.791 | 0.657 - 0.890 | 0,0266 | 85.7 | 75.0 | 79 | 9 | 3.44 | 0.2 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF≤172 pg/mL | 0.854 | 0.723 – 0.938 | 0.0062 | 85.4 | 75.0 | 95 | 50 | 3.44 | 0.2 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + С786С genotype of eNOS gene | 0.820 | 0.710 – 0.901 | 0.0058 | 84.4 | 57.1 | 95 | 29 | 1.95 | 0.28 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + sST2 ≥35 pg/mL | 0.821 | 0.691 – 0.913 | 0.0244 | 87.5 | 80 | 98 | 40 | 4.35 | 0.16 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + С786С genotype of eNOS gene | 0.887 | 0.764 – 0.960 | 0.0026 | 85.0 | 66.7 | 92 | 50 | 2.46 | 0.19 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + sST2 ≥35 pg/mL | 0.880 | 0.722 – 0.966 | 0.0356 | 85.7 | 66.7 | 92 | 50 | 2.61 | 0.21 |
| Val66Met in BDNF gene + MIF≥2792.7 pg/mL + А1166С in ATІІR1 gene + VEGF ≤172 pg/mL + С786С genotype of eNOS gene + sST2 ≥35 pg/mL | 0.913 | 0.765 – 0.982 | 0.0182 | 89.3 | 85.7 | 96 | 63 | 6.36 | 0.13 |
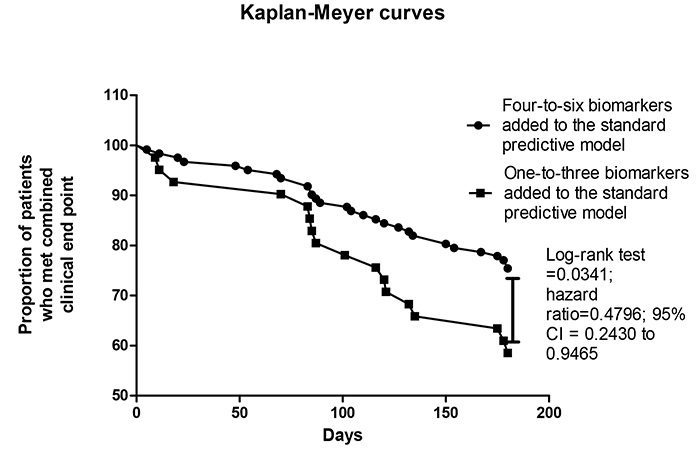
Kaplan-Meyer analysis for endpoint accumulation trends in STEMI patients who were positive for different number of biomarkers added to the standard predictive model
Fig. (4) reports the trend for combined clinical endpoint accumulation in STEMI patients who were positive in four-to-six biomarkers + standard predictive model versus one-to-three biomarkers added to the standard predictive model. The comparison of the endpoint accumulation curves has shown that there were significant differences between patients in accumulation of combined clinical endpoint when they were positive in four and more biomarkers in comparison to three and less biomarkers (Log-rank test =0.0341; hazard ratio=0.4796; 95% CI = 0.2430 to 0.9465).
3.2. Comparison of the Different Predictive Models
We compared predictive values for the standard model and other models based on adding one and more biomarkers to the standard model (Table 6). In fact, there were significant differences between predictive models based on combinations of five and more biomarkers with the standard model and other models including the standard model. When added to the standard model (TIMI STEMI Risk Score+ acute HF Killip class ≥ II + NT-proBNP > 300 pg / mL + troponin >0.05 ng/mL) four biomarkers, the multiple marker risk score significantly improved the C-statistic (area under the curve, 0.883 [95% CI= 0.756-0.958] versus 0.547 [0.450-0.620]; P=0.0028), net reclassification index (0.59; P=0.001), and integrated discrimination index (0.099; P=0.046). Combination of five biomarkers improved standard model to AUC=0.910 (95% CI=0.758 – 0.981; P=0.0294), net reclassification index (0.97; P=0.001), and integrated discrimination index (0.12; P=0.032)
Then we calculated the Wald test value and predictive coefficient for each model and constructed an original score with a modified predictive point suitable for each biomarker (Table 7). For instance, in the standard model, serum levels of VEGF ≤172 pg/mL were allocated one point of predictive score according to the predictive weight based on the estimated predictive coefficient, whereas two pints were allocated for presentation of С786С genotype of eNOS gene, А1166С in ATІІR1 gene and serum levels of sST2 ≥35 pg/mL and MIF ≥2792.7 pg/mL. The total score were reckoned 10 points (Full model -2 Log Likelihood=20.06; Null model -2 Log Likelihood=35.99; P = 0.0434). We hypothesized that the median score (five points) may render patients with different risks in follow-up and that six-to-ten points could discriminate a high risk of a poor clinical outcome; whereas a few numbers of score points might associate with better prognosis.
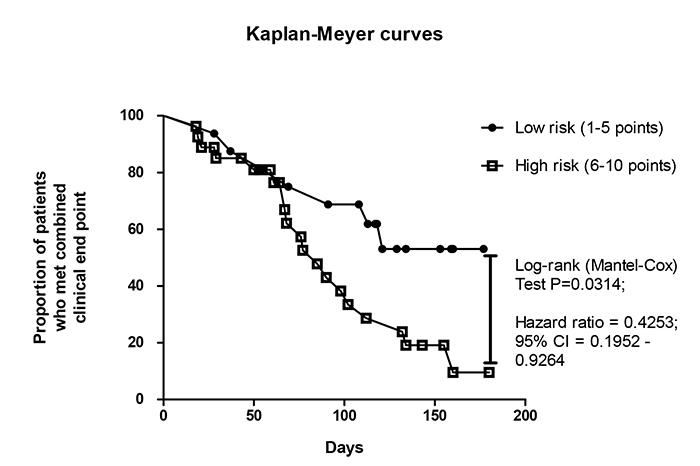
Kaplan-Meyer analysis for endpoint accumulation trends in STEMI patients who were distinguished in total risk score index
Using Kaplan-Meyer analysis, we found that STEMI patients who had six and more score points have demonstrated significantly poor prognosis than individuals with five and less score points (Fig. 5). In fact, the original score system based on biomarker-guided prediction model has appeared better than the standard model in discriminative ability to predict combined clinical outcome in STEMI patients treated with complete revascularization.
| Variables | Depended Variable: Combined End Point | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | NRI | IDI | |||||||
| M | 95% CI | Р value | M | 95% CI | P value | M | 95% CI | P value | |
| Standard model | 0.547 | 0.450 – 0.620 | - | Reference | - | - | Reference | - | - |
| Any single biomarker + Standard model vs. Standard model | 0.664 | 0.541 – 0.773 | 0.1334 | 0.23 | 0.20 – 0.25 | 0.68 | 0.031 | 0.024 – 0.039 | 0.66 |
| Two biomarkers + Standard model vs. Standard model | 0.727 | 0.601 – 0.822 | 0.0496 | 0.30 | 0.21 – 0.42 | 0.72 | 0.033 | 0.022 – 0.045 | 0.88 |
| Three biomarkers + Standard model vs. Standard model | 0.774 | 0.630 – 0.882 | 0.0371 | 0.38 | 0.23 – 0.48 | 0.12 | 0.069 | 0.047 – 0.086 | 0.12 |
| Four biomarkers + Standard model vs. Standard model | 0.883 | 0.756 – 0.958 | 0.0028 | 0.59 | 0.42 – 0.67 | 0.001 | 0.098 | 0.054 – 0.25 | 0.046 |
| Five biomarkers + Standard model vs. Standard model | 0.910 | 0.758 – 0.981 | 0.0294 | 0.97 | 0.60 – 1.38 | 0.001 | 0.12 | 0.090 – 0.24 | 0.032 |
| Predictive Models | AUC | Wald Test | Predictive Coefficient | Predictive Point |
|---|---|---|---|---|
| Standard model | 0.547 | 0.443 | 38.0 | 1 |
| MIF ≥2792.7 pg/mL | 0.664 | 3.4873 | 57.98 | 2 |
| А1166С in ATІІR1 gene | 0.650 | 4.0713 | 64.1 | 2 |
| VEGF ≤172 pg/mL | 0.668 | 4.0473 | 53.5 | 1 |
| С786С genotype of eNOS gene | 0.686 | 5.9783 | 67.5 | 2 |
| sST2 ≥35 pg/mL | 0.706 | 7.1956 | 64.0 | 2 |
| Total | - | - | 345.08 | 10 |
4. DISCUSSION
The results of our study have demonstrated that the original predictive model based on multiple biomarkers may reliably predict poor clinical outcomes in STEMI patients who underwent complete revascularization. In fact, after adjustment for TIMI score, each of the six biomarkers were significantly associated with higher odds of combined clinical outcomes, such as С786С genotype of eNOS gene (rs2070744), Val66Met in BDNF gene (rs6265), А1166С in ATІІR1 gene (rs5186), MIF≥2792.7 pg/mL, VEGF ≤172 pg/mL, sST2 ≥35 pg/mL. We also developed the original risk score that was constructed from these biomarkers being weighted according to their predictive coefficients. As a result, we unleashed the fact that STEMI patients having five and more points of the original predictive score exhibited worse prognosis than those who had lower numbers of score points. Moreover, we did not rule-in the null hypothesis that a new multiple biomarker predictive model would not be better than traditional risk scores including TIMI and its combination with natriuretic peptides, peak levels of troponins and acute HF. In contrast, we first found that the combination of circulating and genetic biomarkers reflecting pathogenesis of cardiac and vascular remodeling strongly predicted poor six-month outcomes after complete PCI among STEMI.
Previous clinical studies have revealed that microvascular occlusion and declined flow reserve after PCI tightly corresponded to post-STEMI regional contractility dysfunction, poor diastolic performances, late Left Ventricular (LV) dilation and reduced LV ejection fraction [30, 31]. There is evidence that adverse cardiac and vascular remodeling mediate MACEs for one year after STEMI even when culprit artery damage and ischemia-related artery stenosis have been recovered and adequate perfusion through large coronary arteries has been completely restored [32]. Interestingly, traditional risk scores, such as TIMI and GRACE, as well as biomarkers of myocardial necrosis (cardiac troponins), inflammation (C-reactive protein, myeloperoxidase, growth/differential factor-15), fibrosis (sST2), biomechanical stress (natriuretic peptides, pro-adrenomedullin) emerged as significant and complementary predictors of MACEs and 30-day mortality for STEMI patients [33-35], while these findings have not been adjusted for a full recovery of blood flow through culprit artery. Although these biomarkers strongly related to myocardial stress, myocyte necrosis, and inflammation and were independently associated with the mortality rate in STEMI patients, there were not sufficient differences in predictive ability between traditional risk scores and biomarker-based scores among complete re-perfused STEMI patients [36]. In contrast, we found that elevated serum levels of several circulating biomarkers, such as MIF≥2792.7 pg/mL and sST2 ≥35 pg/mL, as well as deficiency of circulating pool of VEGF (≤172 pg/mL) were the most reliable complementary predictors for post-STEMI complications. Probably, these three biomarkers are the best fitted to the description of pathogenetic evolution of microvascular occlusion and inadequate distal reperfusion after PCI leading to post-STEMI adverse cardiac remodeling. However, a large body of evidence supports the fact that MIF and VEGF are counteracting factors contributing to vascular integrity and vasculogenesis triggering numerous injury stimuli including ischemia, hypoxia, re-perfused damage [37]. At the same time, sST2 may non-specifically reflect the activity of pro-inflammatory reaction and the impact of immune-relating mechanisms, reactive oxygen species production and enhancing stress-mediated autophagy on vascular integrity and myocardial biomechanical stress and injury [38, 39]. Finally, this combination of the biomarkers appeared to be distinguished from traditionally used natriuretic peptides and myocardial necrosis biomarkers.
Because microvascular inflammation and endothelial dysfunction of small coronary artery accompany to no-reflow phenomenon and are associated well with hibernation and stunning of the myocardium, we hypothesized that SNP of genes playing the pivotal role in inflammation, vascular integrity, reparation, function and angiogenesis, may coordinate susceptibility to myocardial injury in STEMI and thereby mediate CV risk and risk of death. Indeed, previous studies have revealed the association between the endothelial nitric oxide synthase gene T-786C polymorphism and CV mortality, risk of myocardial infarction, HF and re-hospitalization after STEMI [40, 41]. SNP Lys198Asn in EDN-1 gene (rs5370) and SNP Т344С in CYP11B2 gene (rs1799998) corresponded to a risk of CV disease, atrial fibrillation and HF [42, 43]. SNP Val66Met in BDNF gene (rs6265) was associated with lowered serum BDNF concentration and positively correlated to an increased risk of CV disease and mortality rate [44]. Additionally, STEMI patients and individuals with unstable angina have been reported to have increased serum levels of BDNF in the coronary circulation in comparison with stable angina patients, suggesting that BDNF may detrimentally influence plaque stability [45]. SNP А1166С in ATІІR1 gene was accompanied by overexpressed AT-1 receptors on the surface of endothelial cells and well corresponded to a risk of CV events [46, 47]. Although all these SNPs are involved in the pathogenesis of microvascular dysfunction, just three of them (С786С genotype of eNOS gene, Val66Met in BDNF gene, and А1166С in ATІІR1 gene) appeared to be having a clinical relevance for the prediction of CV events in STEMI patients after PCI. However, four-to-six combinations based on circulating and genetic biomarkers have exhibited well-balanced sensitivity and specificity and were enrolled for further validation. The prognostic discriminatory capacity of our original stratification model was confirmed by Kaplan-Meyer analysis, which unleashed that STEMI patients having 5 points and more of total points for original risk score had significantly worse clinical prognosis compared to those who had lower total score index. Thus, our hypothesis that personifying original risk score allows much better stratification of STEMI patients at risk than traditional TIMI score, our results have shown that GRACE and SYNTAX were not better than standard model based on a combination of TIMI score +acute HF Killip class ≥ II + NT-proBNP > 300 pg / mL + troponin >0.05 ng/mL. In fact, traditional score systems do not allow correct stratifying of STEMI patients after successful complete revascularization. Large clinical studies are required to investigate in detail our findings and carefully validate new predictive scores.
4.1. Study Limitations
This study has some limitations. The first limitation is the low number of patients involved in the investigation. Measuring several biomarkers, for instance, serum BDNF and activity of eNOS, in the study was not feasible and we used evidence provided by other researchers regarding strong associations between various SNPs and serum levels of these biomarkers. The effect size for the SNP was not stronger in multiple predictive models, but combinations of genetic and circulating biomarkers were sufficiently better than either. These facts require to be elucidated further. Probably, it is needed to scrutinize reasons leading to microvascular occlusion and no-reflow phenomenon using gadolinium-enhanced MRI or PET / CT angiography. The authors suppose that these restrictions might have no significant impact on data interpretation.
CONCLUSION
The new original predictive model based on a combination of circulating and genetic biomarkers was better than the standard model in discriminative ability to predict combined clinical outcome in STEMI patients.
AUTHOR CONTRIBUTION STATEMENT
Conception and design: Olga V. Petyunina; writing of the article: Olga V Petyunina, Mykola P. Kopytsya, and Alexander E. Berezin; methodology: Mykola P. Kopytsya and Alexander E. Berezin; collection of clinical data, data curation, and investigation: Olga V. Petyunina and Mykola P. Kopytsya; statistical analysis: Olga V. Petyunina and Alexander E. Berezin; supervision: Alexander E. Berezin; critical revision of the article for intellectual content: Alexander E Berezin; interpretation of the data: Olga V Petyunina, Mykola P. Kopytsya, and Alexander E. Berezin; validation: Alexander E. Berezin; project administration: Olga V. Petyunina.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All procedures performed in the study were in accordance with the ethical standards and approved by the local ethical and deontology committee of GI “L.T.Malaya TNI NAMSU”, Kharkiv, Ukraine (Protocol No. 8).
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures were followed in accordance with the ethical standards of the committee responsible for human experimentation (institutional and national), and with the Helsinki Declaration of 1975, as revised in 2013.
CONSENT FOR PUBLICATION
Written voluntary informed consent was obtained from each patient before entering the study.
AVAILABILITY OF DATA AND MATERIALS
Not applicable.
FUNDING
The study was supported by Government Institution “L.T.Malaya Therapy National Institute NAMSU”, Kharkiv, Ukraine, State Registration No. 0117U003028 / Ukraine.
CONFLICTS OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
There are no previous presentations of the information reported in the article. We thank Dr. Nataliia Tytarenko and Dr. Igor Polivenok for performing ultrasound examination and PCI respectively. Additionally, we thank, Dr. Galina Bugrimenko for her excellent technical assistance. Permission to acknowledge has been obtained.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers website along with the published article.


